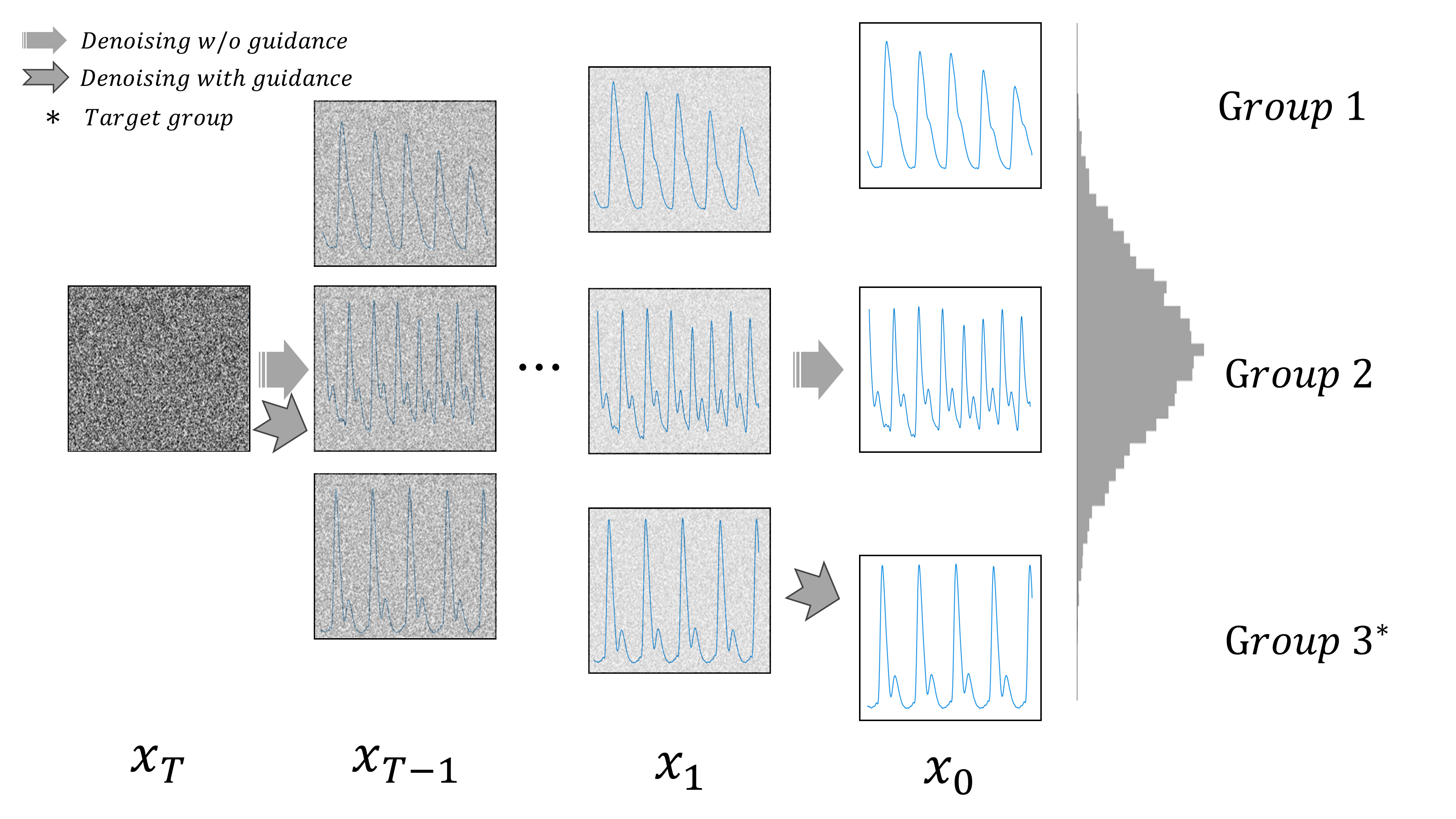
This repository contains PyTorch implemenations of Data Augmentation of PPG Signals Through Guided Diffusion by Python.
In this study, we tackle the challenge of data imbalance in medical datasets by generating high-quality, diverse Photoplethysmogram (PPG) signals using regressor-guided diffusion models. Our approach, a first in this field, effectively enhances Arterial Blood Pressure predictions from PPG data. We address signal noise issues through auxiliary class prediction and diffuse step scheduling, resulting in significant performance improvements in benchmark datasets.
.
├── abp.py
├── data.py
├── denoising_diffusion_pytorch
│ ├── attend.py
│ ├── classifier_free_guidance.py
│ ├── cond_fn.py
│ ├── continuous_time_gaussian_diffusion.py
│ ├── denoising_diffusion_pytorch_1d_guided.py
│ ├── denoising_diffusion_pytorch_1d.py
│ ├── denoising_diffusion_pytorch.py
│ ├── elucidated_diffusion.py
│ ├── fid_evaluation.py
│ ├── guided_diffusion.py
│ ├── __init__.py
│ ├── learned_gaussian_diffusion.py
│ ├── model.py
│ ├── nn.py
│ ├── ppg_model.py
│ ├── resample.py
│ ├── resnet.py
│ ├── simple_diffusion.py
│ ├── version.py
│ ├── v_param_continuous_time_gaussian_diffusion.py
│ └── weighted_objective_gaussian_diffusion.py
├── main.py
├── models.py
├── paths.py
├── README.md
├── reg_resnet.py
├── setup.py
├── sh
│ ├── sample_diffusion.sh
│ ├── train_and_sample_diffusion_for_ppgbp.sh
│ ├── train_and_sample_diffusion_for_sensors.sh
│ └── train_reg_res.sh
└── utils.py
Our repository involves a three-step process:
- Training the guidance regressor
- Training the diffusion model
- Sampling target PPG signals using the trained guidance regressor and diffusion model.
For this, we utilize reg_resnet.py and main.py. reg_resnet.py is for training the guidance regressor, and main.py manages both the training and sampling of the diffusion model. If the diffusion model has already been trained, you can use the --sample_only flag in main.py to skip the training phase and proceed directly to sampling.
Also, for hyperparameter sweeping, we used files located in ./sh .
We used the following Python packages for core development. We tested on Python 3.11.4.
pytorch 1.9.0
pandas 1.3.3
numpy 1.22.4
scikit-learn 0.24.2
scipy 1.7.1
This script is used for generating PPG (Photoplethysmography) data with regressor guidance.
--seq_length: Sequence length. Default is625(625 for BCG and sensors, 262 for PPG-BP).--train_batch_size: Training batch size. Default is32.--min_max: If set toFalse, disables Min-Max normalization of data. Default isTrue.--benchmark: Specifies the benchmark dataset. Default is'bcg'.--train_fold: Specifies the train fold. for cross validation setting of BP benchmark.--channels: Number of channels. Default is1.
--disable_guidance: If set, disables the use of guidance. Default isFalse.--reg_train_loss: Specifies the regression training loss. Default is'group_average_loss'.--reg_selection_dataset: Dataset for regression selection. Default is'val'.--reg_selection_loss: Loss function for regression selection. Choices are["erm", "gal", "worst"]. Default is'gal'.
--diffusion_time_steps: Number of diffusion time steps. Default is2000.--train_num_steps: Number of training steps. Default is32.--train_lr: Learning rate for training. Default is8e-5.--optim: Optimization algorithm. Default is'adam'.--dropout: Dropout rate. Default is0.
--sample_only: If set, stops training and enables only sampling. Default isFalse.--sample_batch_size: Batch size for sampling. Default isNone.--target_group: Target group for sampling. Choices are[-1, 0, 1, 2, 3, 4], where-1is all,0is hyp0,1is normal,2is perhyper,3is hyper2,4is crisis. Default is-1(all).--t_scheduling: Scheduling type fort. Choices are["loss-second-moment", "uniform", "train-step"]. Default is"uniform".--regressor_scale: Scale of the regressor. Default is1.0.--regressor_epoch: Number of epochs for the regressor. Default is2000.--gen_size: Size for generation. Default is8096.
This script is used for training a guidance regressor for do regression task of noisy PPG data.
--seq_length: Sequence length. Default is625.--train_batch_size: Training batch size. Default is32.--min_max: If set toFalse, disables Min-Max normalization of data. Default isTrue.--benchmark: Specifies the benchmark dataset. Default is'bcg'.--train_fold: Specifies the training fold. Default is0.
--final_layers: Number of final layers in the model. Default is3.--time_linear: If set, uses a linear layer instead of MLP for time embedding. Default isFalse.--auxilary_classification: If set, adds classification as an auxiliary task. Default isFalse.--is_se: If set, uses SE architecture. Default isFalse.--do_rate: Dropout rate. Default is0.5.
--diffusion_time_steps: Number of diffusion time steps. Default is2000.--train_epochs: Number of training epochs. Default is2000.--init_lr: Initial learning rate. Default is0.0001.--weight_decay: Weight decay factor. Default is1e-4.--init_bias: Initial bias. Default is0.2.--final_bias: Final bias. Default is1.--loss: Loss function. Choices are["ERM", "group_average_loss"]. Default is'group_average_loss'.--t_scheduling: Scheduling type fort. Choices are["loss-second-moment", "uniform", "train-step"]. Default is"train-step".--T_max: Maximum value forT. Default is2000.--eta_min: Minimum eta value. Default is0.
The simplest way to use our model is as follows:
# Train Regressor
python reg_resnet.py --lr_init=1e-4 --wd=1e-3 --t_scheduling "train-step" --loss "group_average_loss" --train_fold 0 --auxiliary
# Train and sample PPG signal with diffusion model
python main.py --reg_train_loss "group_average_loss" --train_num_steps 32 --train_lr 8e-05 --train_fold 0 --reg_selection_loss "gal"
Below are examples of t-SNE visualizations generated by the script. These visualizations make it easier to understand that the data generated by the dataset closely matches the group distribution of the actual data.
| Hypo Group | Normal Group |
|---|---|
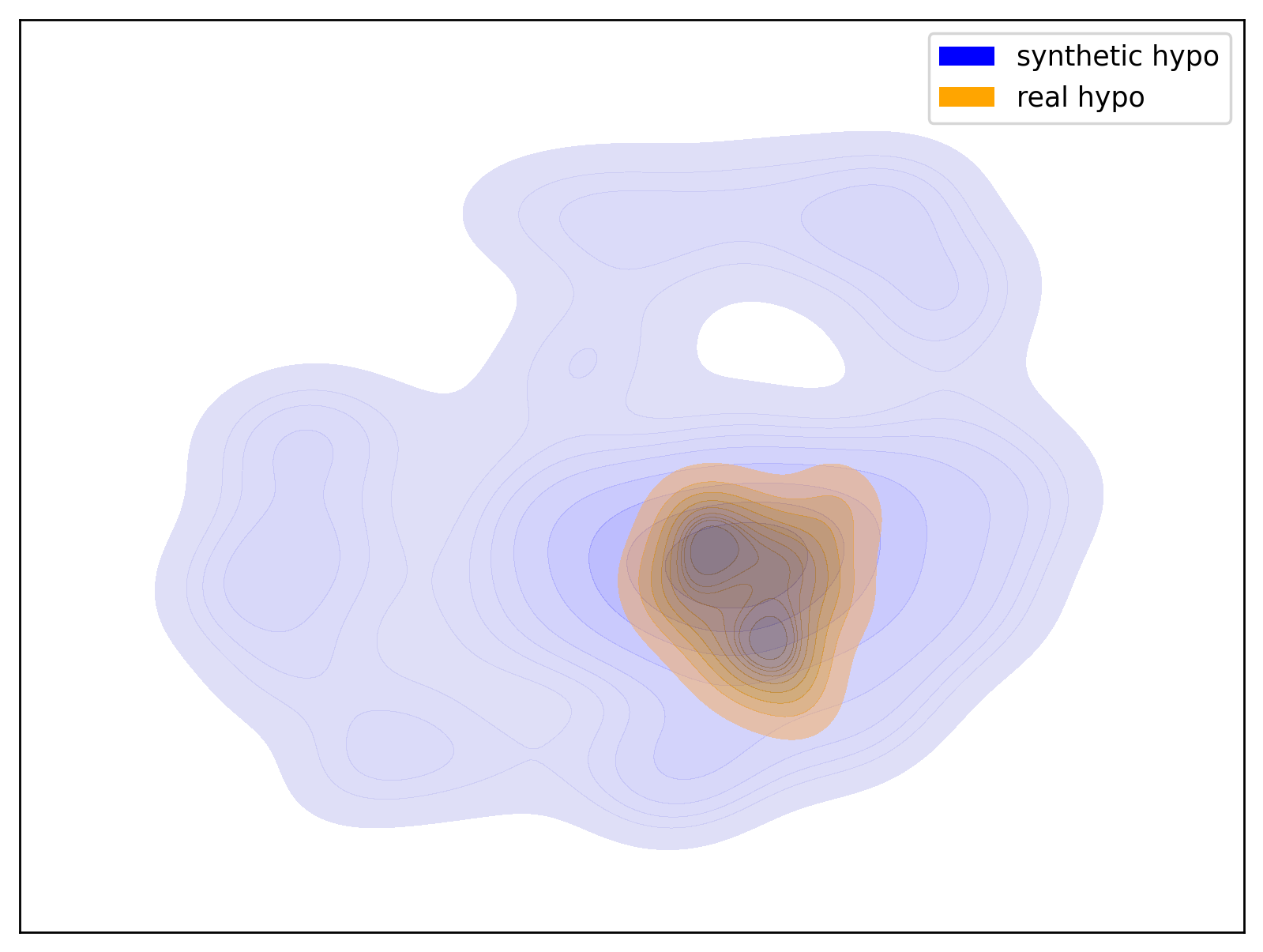 |
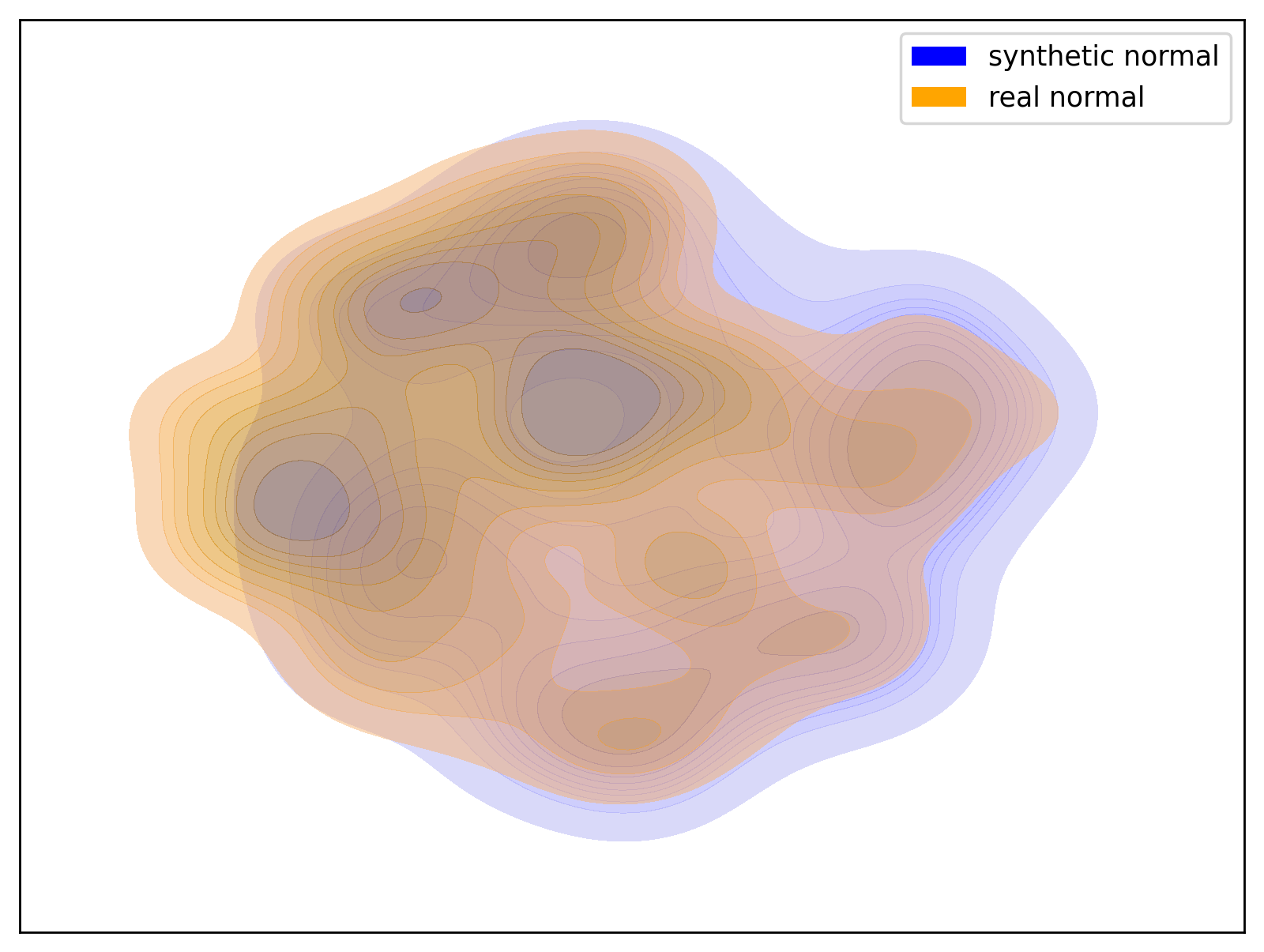 |
| t-SNE visualization of the Hypo group | t-SNE visualization of the Normal group |
| Prehyper Group | Hyper2 Group |
|---|---|
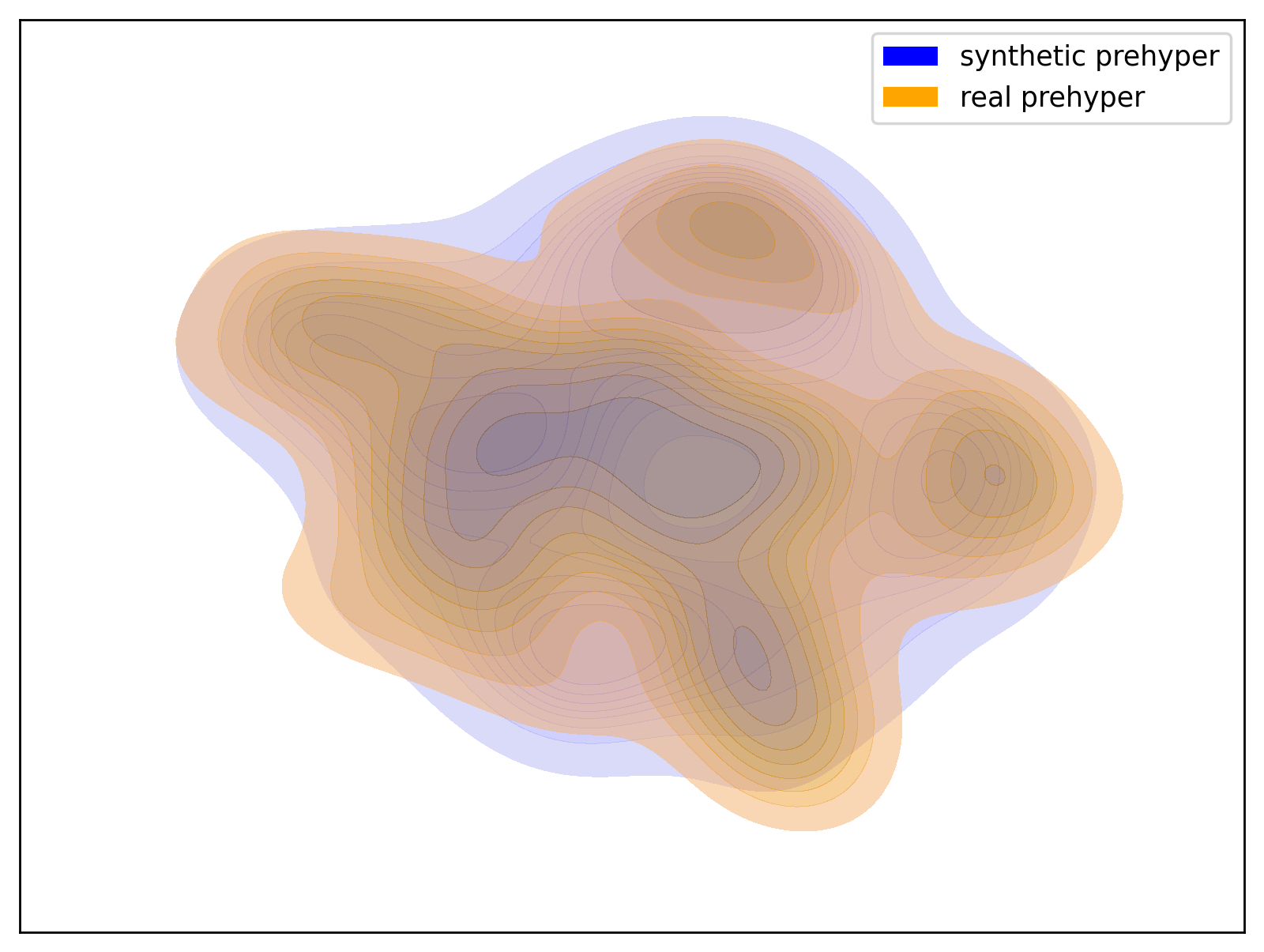 |
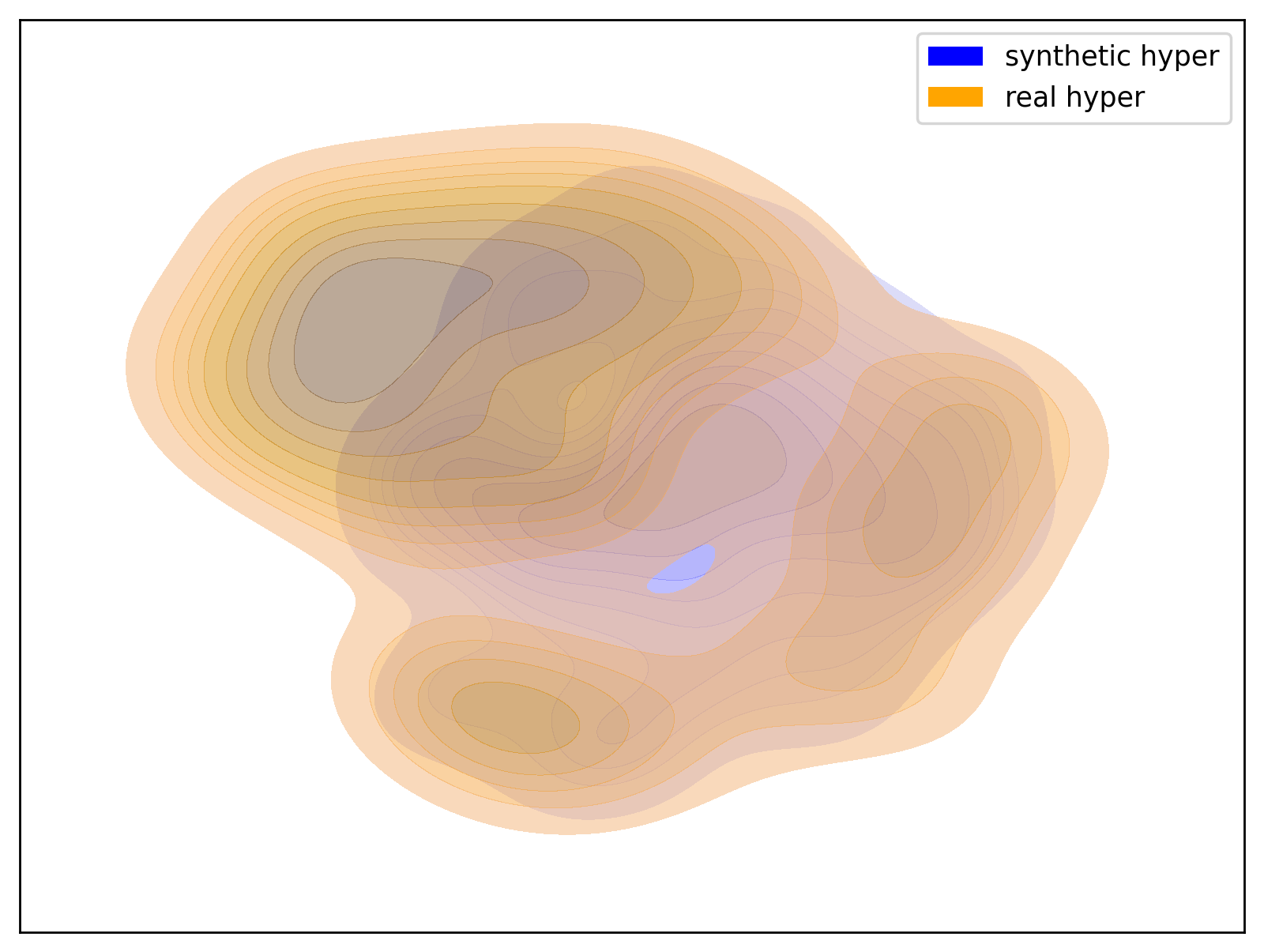 |
| t-SNE visualization of the Prehyper group | t-SNE visualization of the Hyper2 group |