By *Hogeon Seo, *Seunghyeok Back, *Seongju Lee, Deokhwan Park, Tae Kim, and Kyoobin Lee (*: Equal Contribution)
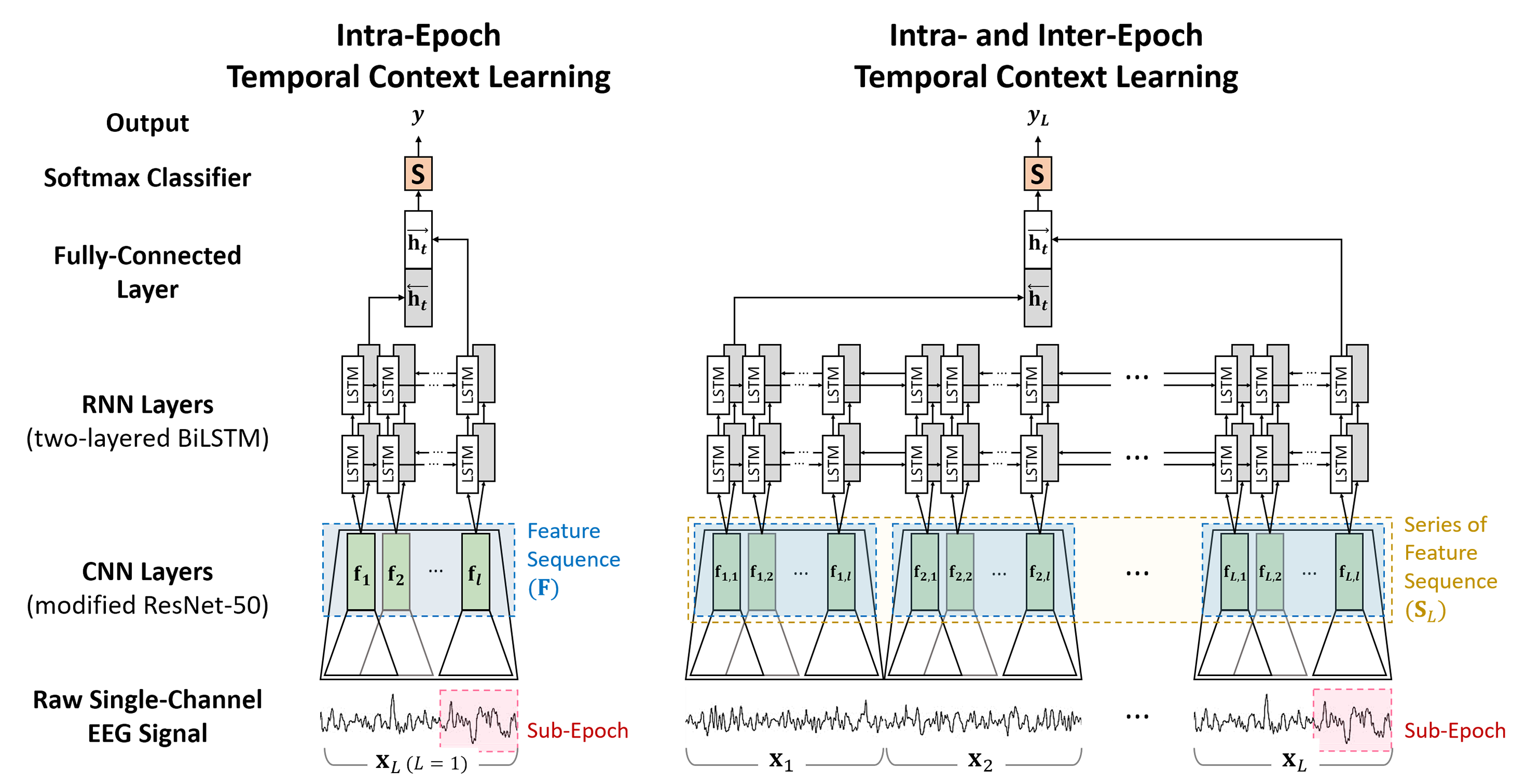
This repo is the official implementation of "Intra- and Inter-epoch Temporal Context Network (IITNet) using Sub-epoch Features for Automatic Sleep Scoring on Raw Single-channel EEG", Biomedical Signal Processing and Control 61 (2020): 102037. [paper]
- (2022.03.22) IITNet official repository is released.
- (2022.0X.XX) DeepSleepNet [1] baseline is added.
- python >=3.7.0
- pytorch >= 1.7.0 (or compatible version to your develop env)
- numpy
- scikit-learn
- pandas
- mne
- terminaltables
- termcolor
- Install PyTorch with compatible version to your develop env from PyTorch official website.
- Install remaining libraries using the following command.
pip install -r requirements.txt
We evaluated our IITNet with MASS, SHHS, and Sleep-EDF dataset. You have to convert dataset from .edf into .npz format.
First, download .edf files and annotations MASS, SHHS, Sleep-EDF. For the MASS and SHHS dataset, you have to request for a permission to access their dataset.
You can download SC subjects of Sleep-EDF using the following commands.
cd data
chmod +x download_physionet.sh
./download_physionet.sh
Second, preprocess .edf into .npz format using the following command. We referred prepare_physionet.py in DeepSleepNet repository.
python prepare_physionet.py
After preprocessing, the hierarchy of ./datasets/ directory will be the following:
./datasets/
├── MASS/
│ └── F4-EOG/
│ ├── 01-03-0001-PSG.npz
│ ├── ...
│ └── 01-03-0064-PSG.npz
├── SHHS/
│ └── C4-A1/
│ ├── shhs1-200001.npz
│ ├── ...
│ └── shhs1-205804.npz
└── Sleep-EDF/
└── Fpz-Cz/
├── SC4001E0.npz
├── ...
└── SC4192E0.npz
Each .npz file contains input eeg epochs with the shape of (total_num_epochs, 30 * sampling_rate) and target labels with the shape of (total_num_epochs) with the key 'x' and 'y', respectively.
You can simply train and evaluate IITNet using just main.py.
$ python main.py --config $CONFIG_PATH --gpu $GPU_IDs
For evaluation only, add --test-only argument when you run the script
- Train and Evaluation MASS (
L=1) using single GPU (gpu_id=0)
$ python main.py --config ./configs/IITNet_MASS_SL-01.json --gpu 0
- Train and Evaluation Sleep-EDF (
L=10) using multiple GPUs (gpu_id=1,2)
$ python main.py --config ./configs/IITNet_Sleep-EDF_SL-10.json --gpu 1,2
- Evalution trained SHHS (
L=10) using single GPU (gpu_id=3)
$ python main.py --config ./configs/IITNet_SHHS_SL-10.json --gpu 3 --test-only
- For each fold, checkpoints that have the best validation loss are saved at
./checkpoints/CONFIG_NAME/. - Overall result is written in
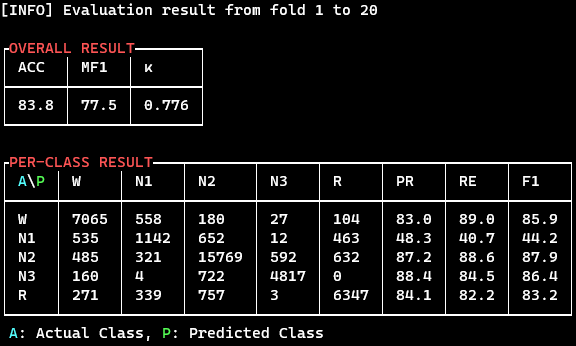
./results/CONFIG_NAME.txtwith the order ofFOLD ACC MF1 KAPPA W-F1 N1-F1 N2-F1 N3-F1 REM-F1. - Overall and per-class results are printed like the following figures:
If you find this project useful, we would be grateful if you cite our work as follows:
@article{seo2020intra,
title={Intra-and inter-epoch temporal context network (IITNet) using sub-epoch features for automatic sleep scoring on raw single-channel EEG},
author={Seo, Hogeon and Back, Seunghyeok and Lee, Seongju and Park, Deokhwan and Kim, Tae and Lee, Kyoobin},
journal={Biomedical Signal Processing and Control},
volume={61},
pages={102037},
year={2020},
publisher={Elsevier}
}
This work was supported by the Institute of Integrated Technology (IIT) Research Project through a grant provided by Gwangju Institute of Science and Technology (GIST) in 2019 (Project Code: GK11470).
[1] A. Supratak, H. Dong, C. Wu, and Y. Guo, “DeepSleepNet: A model for automatic sleep stage scoring based on raw single-channel EEG,” IEEE Trans. Neural Syst. Rehabil. Eng., vol. 25, no. 11, pp. 1998–2008, 2017.
MIT licence