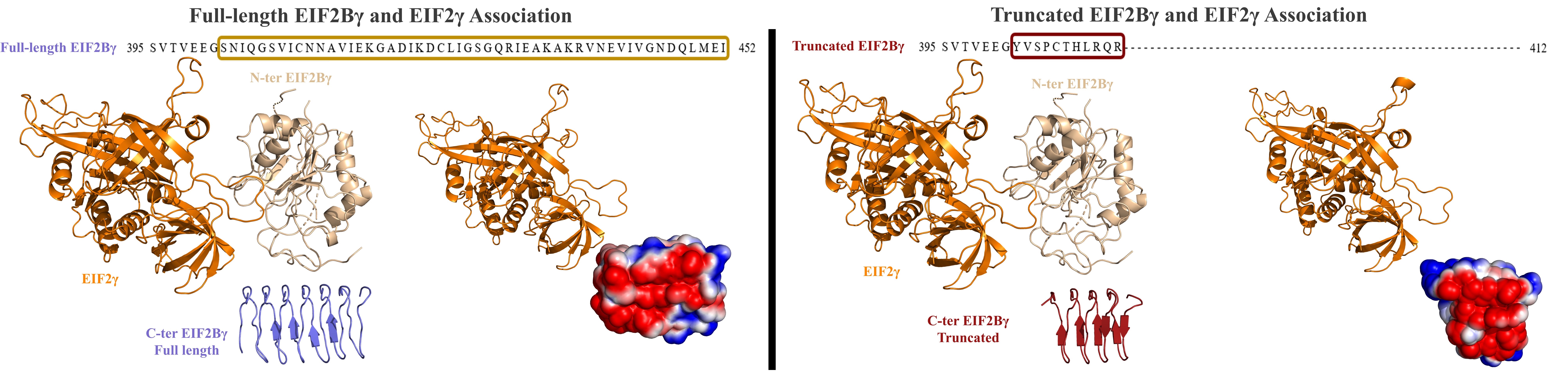
This work presents the structural part of the "A C-term truncated EIF2Bγ protein encoded by an intronically polyadenylated isoform introduces unfavorable EIF2Bγ-EIF2γ interactions" study by Ayca Circir, Gozde Koksal Bicakci, Busra Savas, Didem Naz Doken, Onur Henden, Tolga Can, Ezgi Karaca, Ayse Elif Erson-Bensan.
The main subject of this study is EIF2Bγ which is a member of EIF2B complex regulating the exchange of GDP (produced by GTP hydrolysis during Met-tRNAi and EIF2 association) to GTP. In 2019, a new arrangement of EIF2:EIF2B is resolved (Kashiwagi K. et al., 2019). In 6K71 PDB structure, EIF2γ interacts with EIF2Bε forming an active complex which is capable to perform nucleotide exchange activity. But in 6K72 structure, EIF2γ interacts with EIF2Bγ in a way that nucleotide exchange activity is precluded. In inactive mode of EIF2:EIF2B complex, EIF2γ interacts with both N and C terminals of EIF2Bγ. Here, we investigated the effect of an intronically polyadenylated EIF2Bγ transcript causing C-terminal truncated isoform and full-length (canonical) isoform. For that purpose, we modelled the C terminals of both isoforms with various modeling servers. After selection of the best models according to our goals, we performed standard refinement protocol to enhance EIF2γ:EIF2Bγ interface with HADDOCK2.2. We also calculated the electrostatics potential maps of each complex with PDB2PQR server.
- Models: contains the PDB files of I-patch domain of both isoforms modelled with I-TASSER, iDrug, TrRosetta, Ab-initio Rosetta and QUARK servers.
- Initial_structures: contains all the relevant input files for HADDOCK runs.
- HADDOCK_runs: contains the HADDOCK output files of truncated and full-length isoforms.
- PQR_files: contains the input (PDB) and output (PQR) files obtained by PDB2PQR server.
- Graph: contains the electrostatics energy per residue files obtained by HADDOCK, scripts to obtained the electrostatics energies of the best four complexes, excel file including desired values, graph, jupyter notebook script to obtain the graph along with data file.
git clone https://github.com/CSB-KaracaLab/eif2g-eif2bg-ints.git
or if you would like to get the content directly via wget:
wget https://github.com/CSB-KaracaLab/eif2g-eif2bg-ints/archive/master.zip
Kashiwagi K, Yokoyama T, Nishimoto M, Takahashi M, Sakamoto A, Yonemochi M et al. Structural basis for eIF2B inhibition in integrated stress response. Science (80- ) 2019; 364: 495–499.
This research is supported by EMBO installation grant( (#4421).