phasing and Allele Specific Expression from RNA-seq
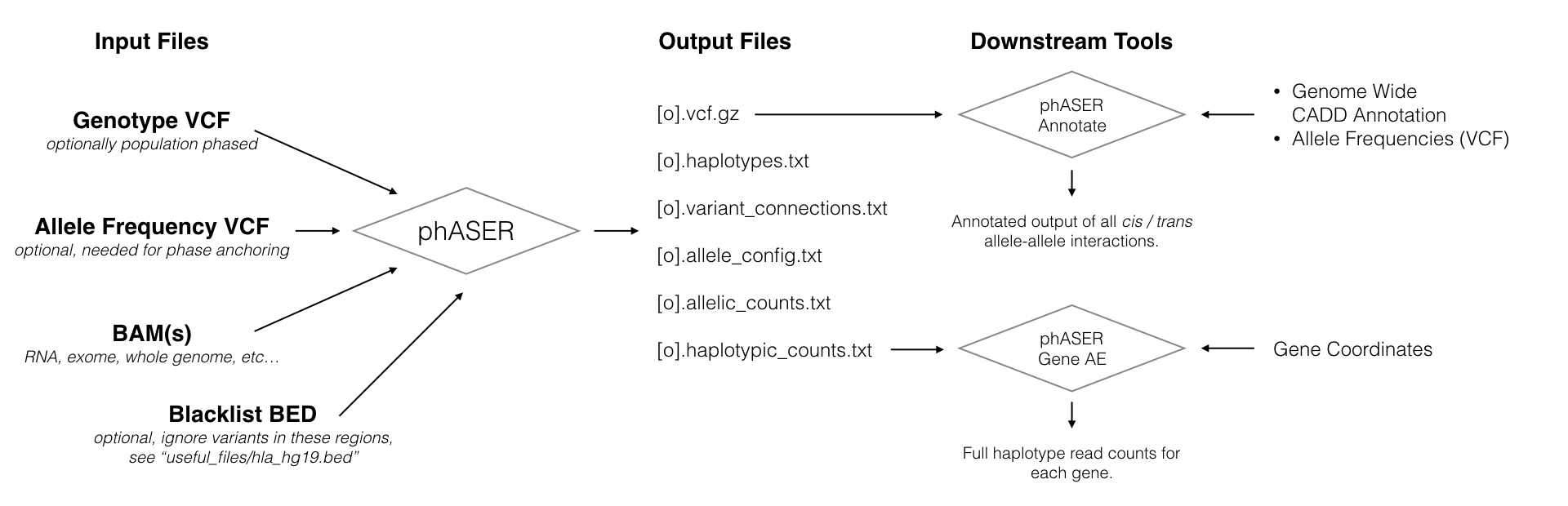
Performs haplotype phasing using read alignments in BAM format from both DNA and RNA based assays, and provides measures of haplotypic expression for RNA based assays.
Developed by Stephane E. Castel in the Lappalainen Lab at the New York Genome Center and Columbia University Department of Systems Biology.
Please see our paper in Nature Communications for details and benchmarking of the method and our publication in Genome Biology for its application to the GTEx v8 data set and details of the phASER-pop extension.
Haplotype-level ASE data is publicly available for all 15,253 samples spanning 54 human tissues from the GTEx project version 8 release through the GTEx Portal under the "Haplotype Expression Matrices" section.
phASER is made available under the GNU GENERAL PUBLIC LICENSE v3.
Documentation Pages: phASER, phASER Annotate, phASER Gene AE, phaser-POP
A bug was introduced in version 0.9.8 (12/16/16) and fixed in version 0.9.9.4 (06/21/17) that caused problems with haplotypic counts when using the --haplo_count_blacklist argument. This bug affects the haplotypic counts generated (haplotypic_counts.txt), and any downstream analyses of those counts, including generating gene level haplotypic expression with phaser_gene_ae. If the --haplo_count_blacklist argument was not specified, then the results were not affected. In addition, a new "hg19_haplo_count_blacklist.bed.gz" file has been uploaded, which addresses problems related to this issue. If you used the --haplo_count_blacklist argument with a version of phASER between 0.9.8 and 0.9.9.3 you must re-run your analyses with version 0.9.9.4+.