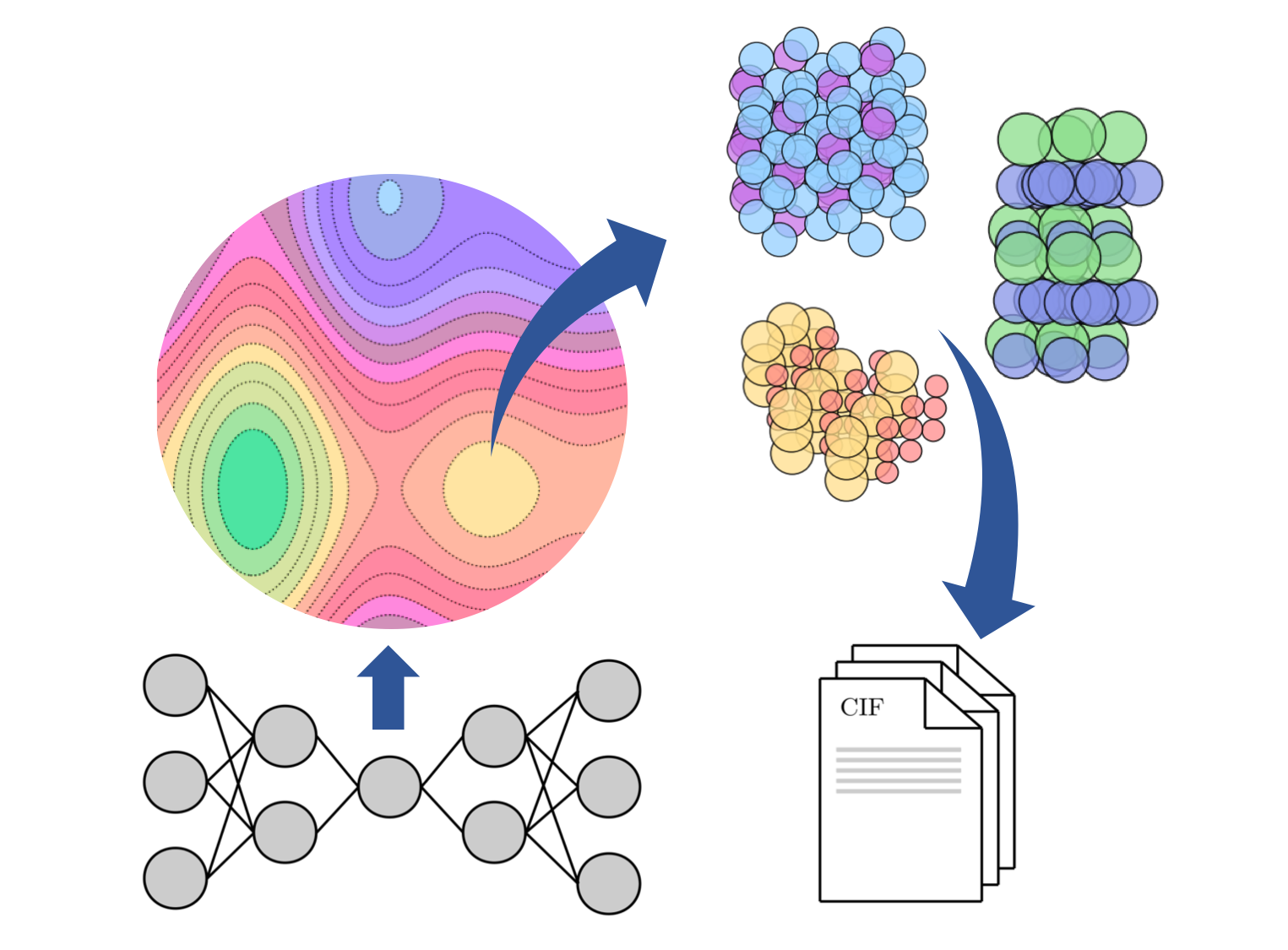
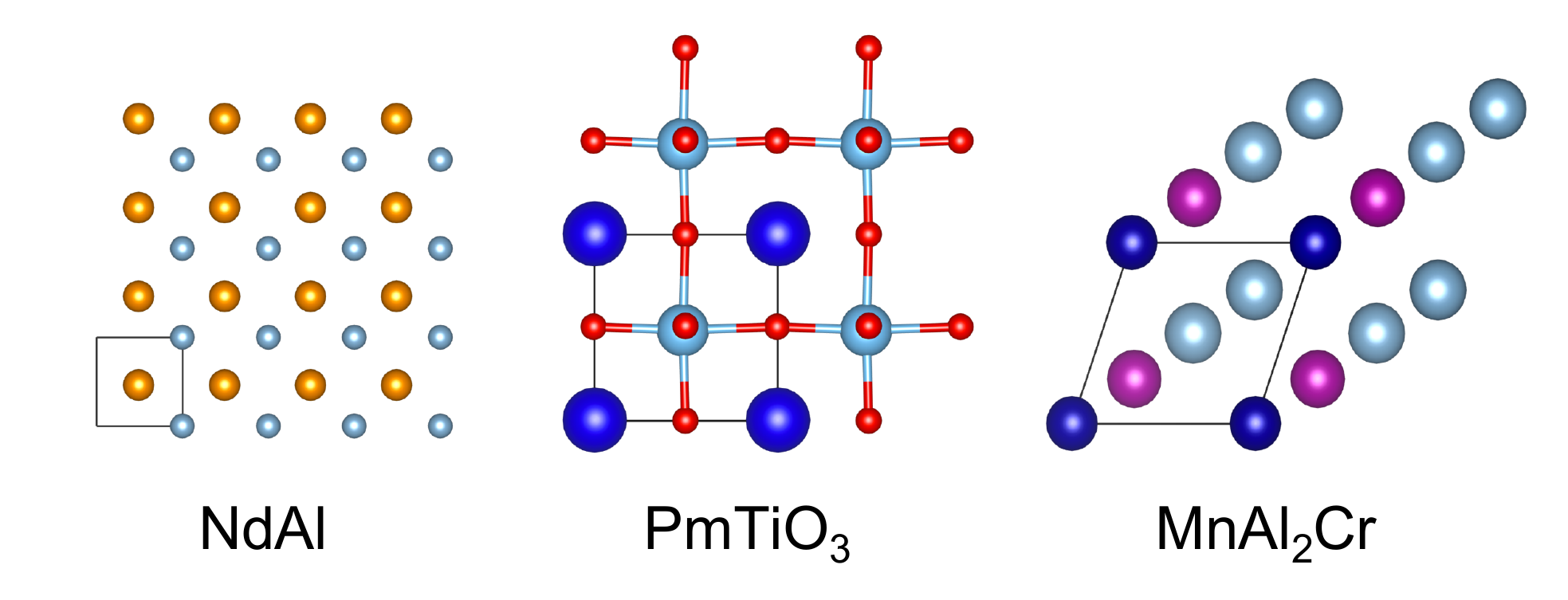
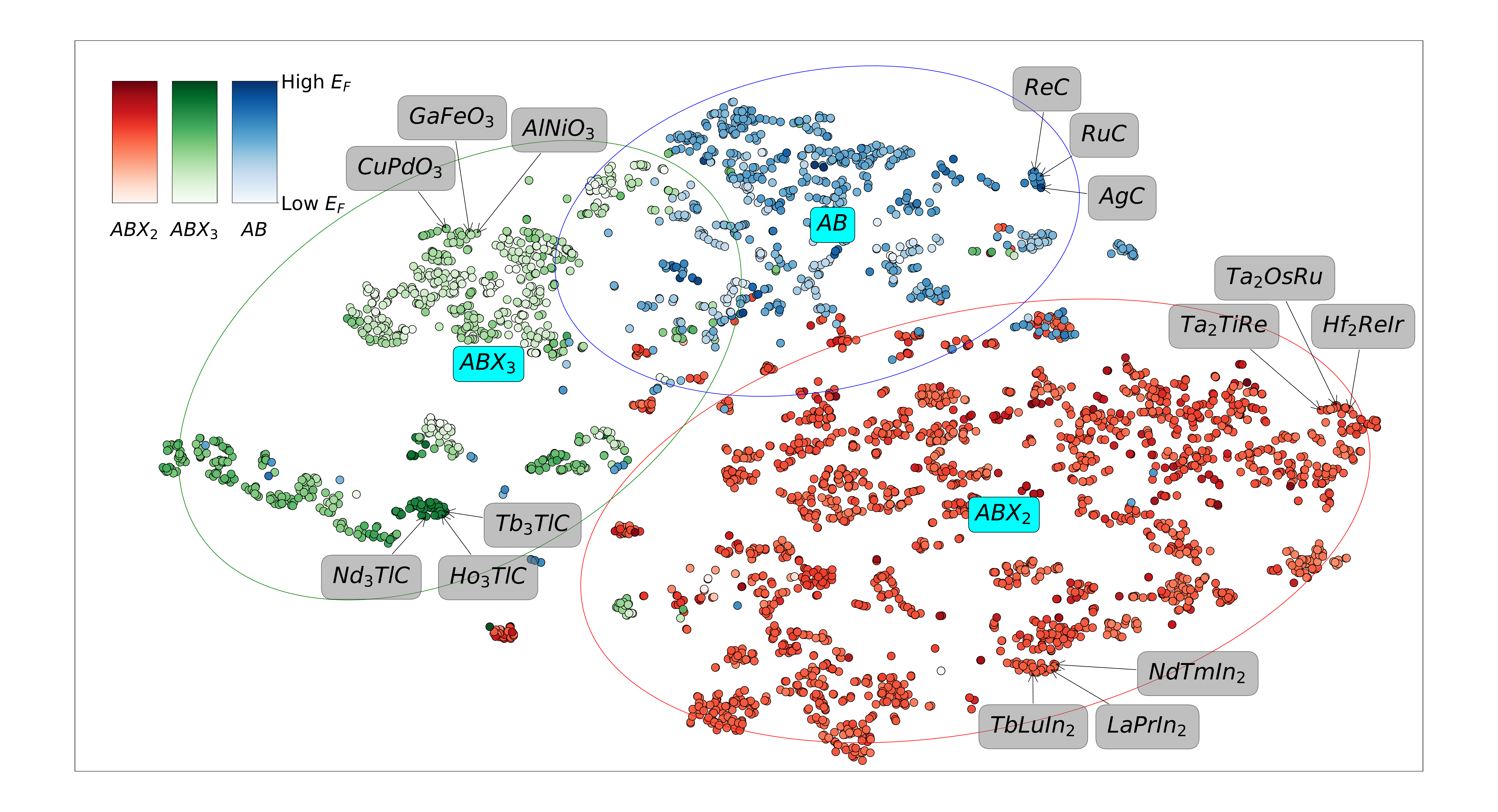
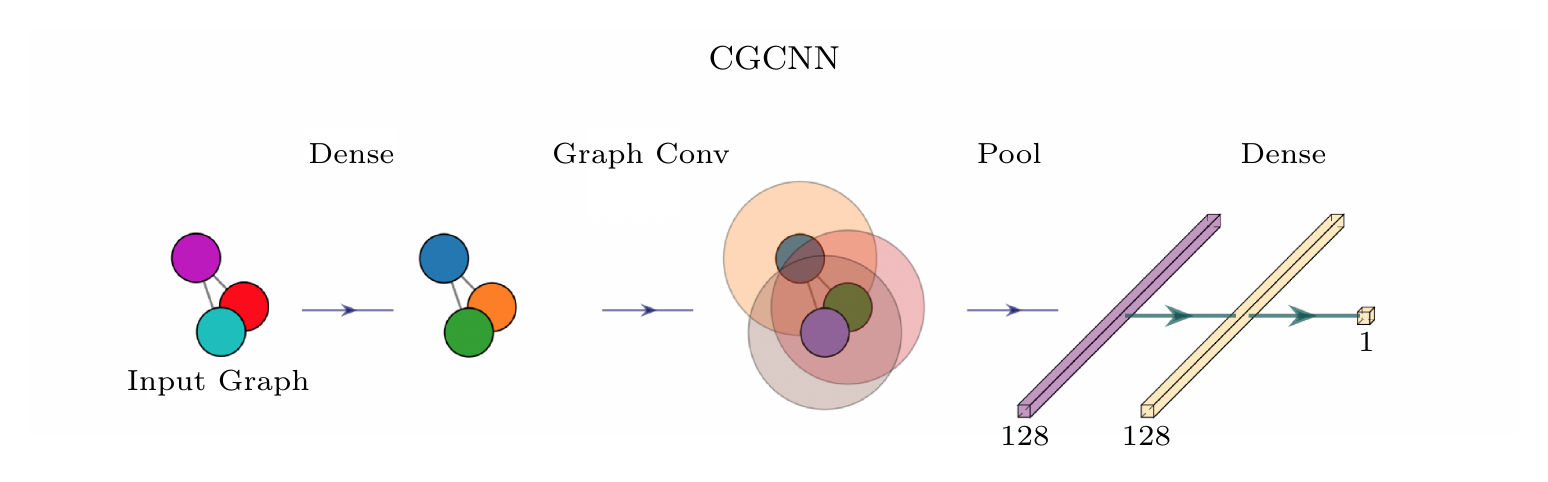
All source code and images are associated with the paper:
C. J. Court, B. Yildirim, A. Jain, J. M. Cole,
"3-D Inorganic Crystal Structure Generation and Property Prediction via Representation Learning",
J. Chem. Inf Model. (accepted for publication) (2020).
Paper HTML
Our pipeline consists of 3 components.
- A Conditional Deep Feature Consistent Variational Autoencoder
- A UNet semantic segmentation network
- A Crystal Graph Neural Network
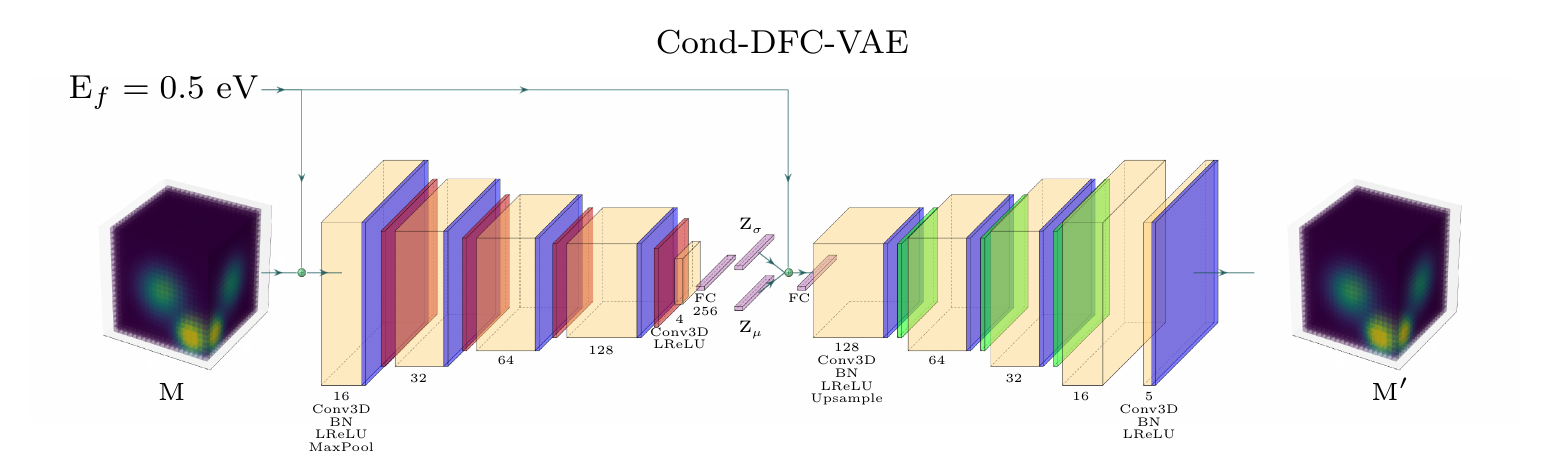 Encoder: 4x 3D convolutions, BatchNorm, ReLU and Maxpooling
Encoder: 4x 3D convolutions, BatchNorm, ReLU and Maxpooling
Bottleneck: 3D convolution, LeakyReLU, Dense (256), 2x Dense (256) ( and
)
Decoder: 4x 3D convolutions, BatchNorm, ReLU and upsampling
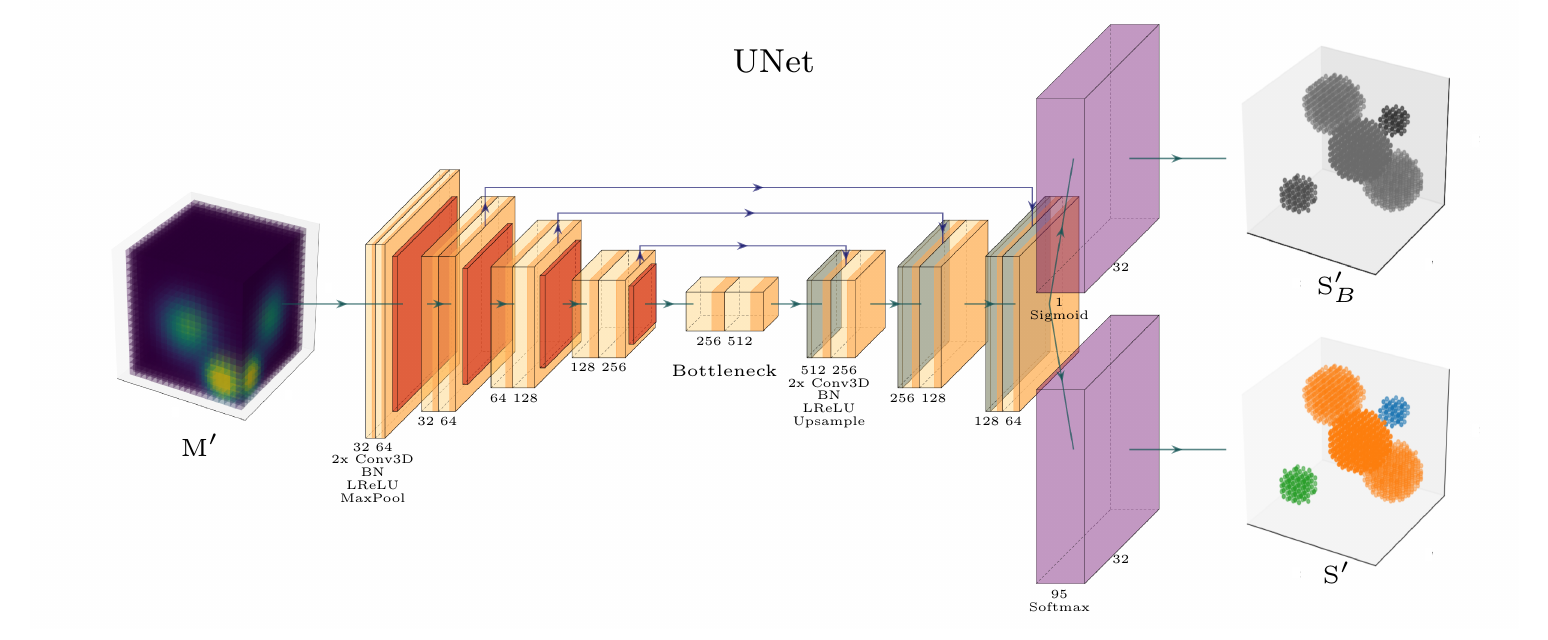 Downward: 4 x 2 x 3D convolutions, ReLU, BatchNorm, and pooling
Downward: 4 x 2 x 3D convolutions, ReLU, BatchNorm, and pooling
Bottleneck: 2 x 3D convolutions, ReLU BatchNorm
Upward: 4 x 2 x 3D convolutions, ReLU, BatchNorm and UpSampling
- Clone the git repository
git clone https://github.com/by256/icsg3d
- Install requirements
python3 -m pip install -r requirements.txt
Tensorflow2.0 is not currently supported, tested with tensorflow-gpu==2.1.0 and Keras==2.3.1 Requires keras-contrib
The system works on crystallographic information files (CIFs) to train the deep learning pipeline. In theory these can be from any source, but by default we use the materialsproject API.
For example, to retrieve all CIFs for cubic perovskites (ABX3):
python3 query_matproj.py --anonymous_formula="{'A': 1.0, 'B': 1.0, 'C':3.0}" --system=cubic --name=perovskites
This will create a data/perovskites folder containing the cifs and a csv with associated properties
The various network input matrices can be created by
mpiexec -n 4 python3 create_matrices.py --name=perovskites
Train the unet for as many epochs as needed
python3 train_unet.py --name perovskites --samples 1000 --epochs 50
Make sure you train the VAE second (as it uses the unet as a DFC perceptual model)
python3 train_vae.py --name perovskites --samples 1000 --epochs 250
- Interpolations in vae latent space
python3 interpolate.py --name perovskites
- Whole pipeline plots
python3 view_results.py --name perovskites
- Evaluate coordinates and lattice params
python3 eval.py --name perovskites
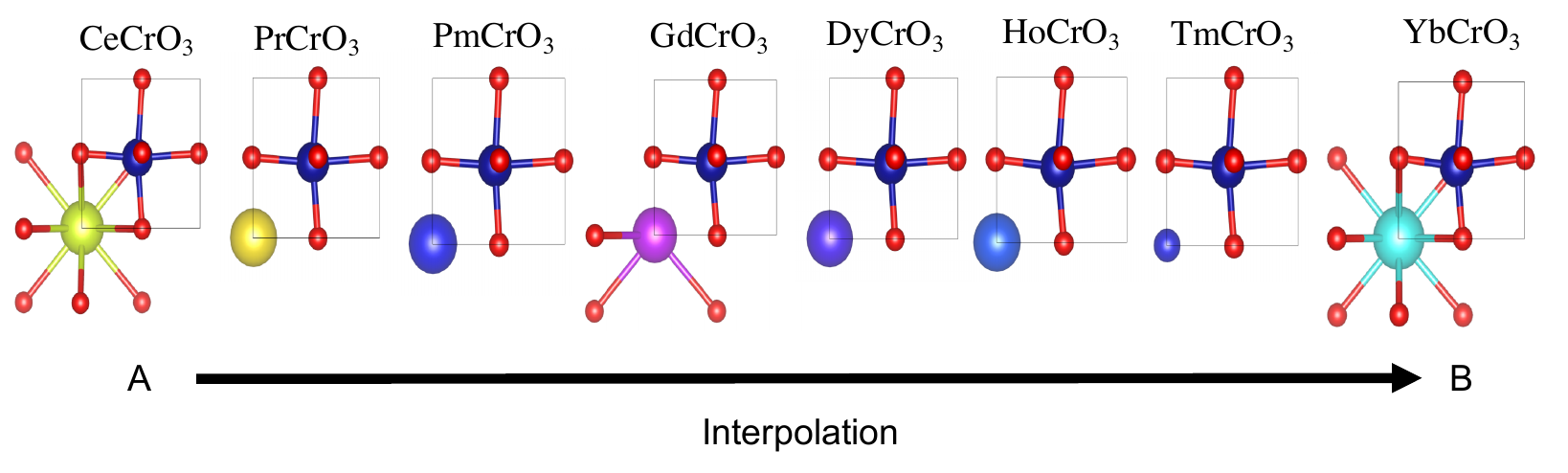
Attempt to generate 1000 new samples arund a base compound CeCrO3 with variance 0.5
python3 generate.py --name perovskites --nsamples 1000 --base CeCrO3 --var 0.5
This will create a new directory where you will find Cifs, density matrices, species matrices and properties for all generated compounds.
The pre-trained models in the models directory. To use these, you'll need to move/rename the models to a corresponding directories:
saved_models/unet/mode/unet_weights_mode.h5 saved_models/unet/mode/class_weights.npy saved_models/vae/mode/vae_weights_mode.hdf5 saved_models/cgcnn/
where mode is the name of your data directory.
C. J. Court, B. Yildirim, A. Jain, J. M. Cole, "3-D Inorganic Crystal Structure Generation and Property Prediction via Representation Learning", J. Chem. Inf. Model. 2020 (accepted for publication).
This project was financially supported by the Engineering and Physical Sciences Research Council (EPSRC, EP/L015552/1), Science and Technology Facilities Council (STFC) and the Royal Academy of Engineering (RCSRF1819\7\10).