Target-driven machine-learning-enabled virtual screening is a machine learning tool developed to accelerate the early-stage hit identification. In this repository, you can find all you need to launch virtual screening against your target protein with different types of input information:
- Mimimum info: a single uniport ID of your target protein (starting point 1);
- Intermediate info: Compound datasets containing active and inactive molecules against homologies of your target of interest (starting point 2);
- Advanced info: Compound datasets containing active molecules directly against your target of interest (starting point 3).
For more information, please refer to our paper (Yuemin Bian): https://www.frontiersin.org/articles/10.3389/fmolb.2023.1163536/full
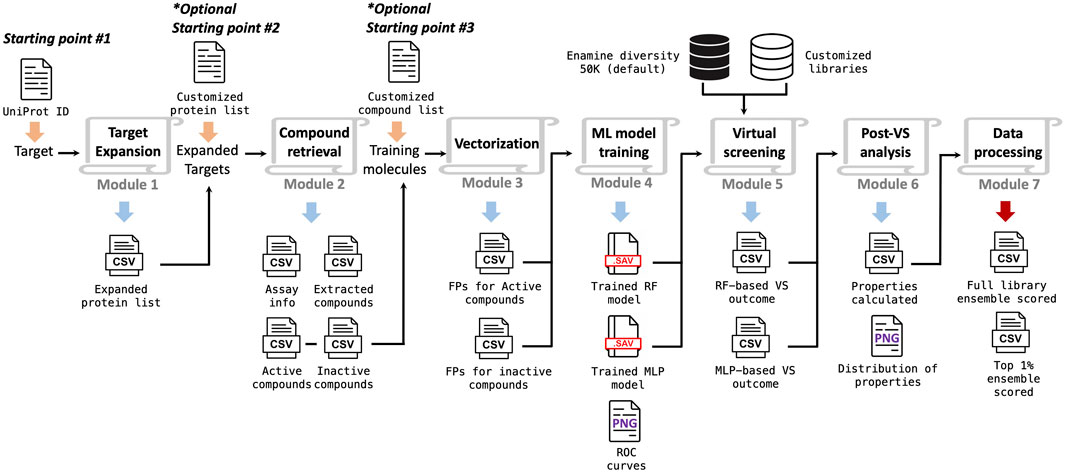
The Starting_point[1-3].sh are the production scripts for launching vritual screening with different types of inputs. Clone the entire repository to your local machine prior to start.
To run the virtual screening, you need to install miniconda.
Then you need to
- Setup a conda virtual environment with python (3.7) and activate it
conda create -n TAME_VS python=3.7
conda activate TAME_VS- Install all third-party packages
pip install -r requirements.txtTo use the included Enamine 50k compound library for final ML virtual screening, please run the followig command from 5_Virtural_screening
python Library_preparation.py -i Enamine_diversity_50K.csv -s 1 -c 2 -f Enamine_diversity_50K_morgan_1024_FPIn March 2024, we added a convert.py file to module 5_Virtual_screening. This convert.py can be used to convert a customized .sdf chemical library into the .csv format. Then the Library_preparation.py should be able to be used for fingerprints calculations.
The following example uses starting point 1 as an example.
- Search for the uniport ID of your target proteins (e.g. P09238);
- Launch the
Starting_point_1.shscript and provide uniport ID and working directory;
bash Starting_point_1.sh