The approach taken to complete this task consisted in three stages:
1.- Load and exploration. 2.- Artifact removal. 3.- Spectral Classification.
All the code discussed here was developed to complete this task with minimal invokation of toolboxes.
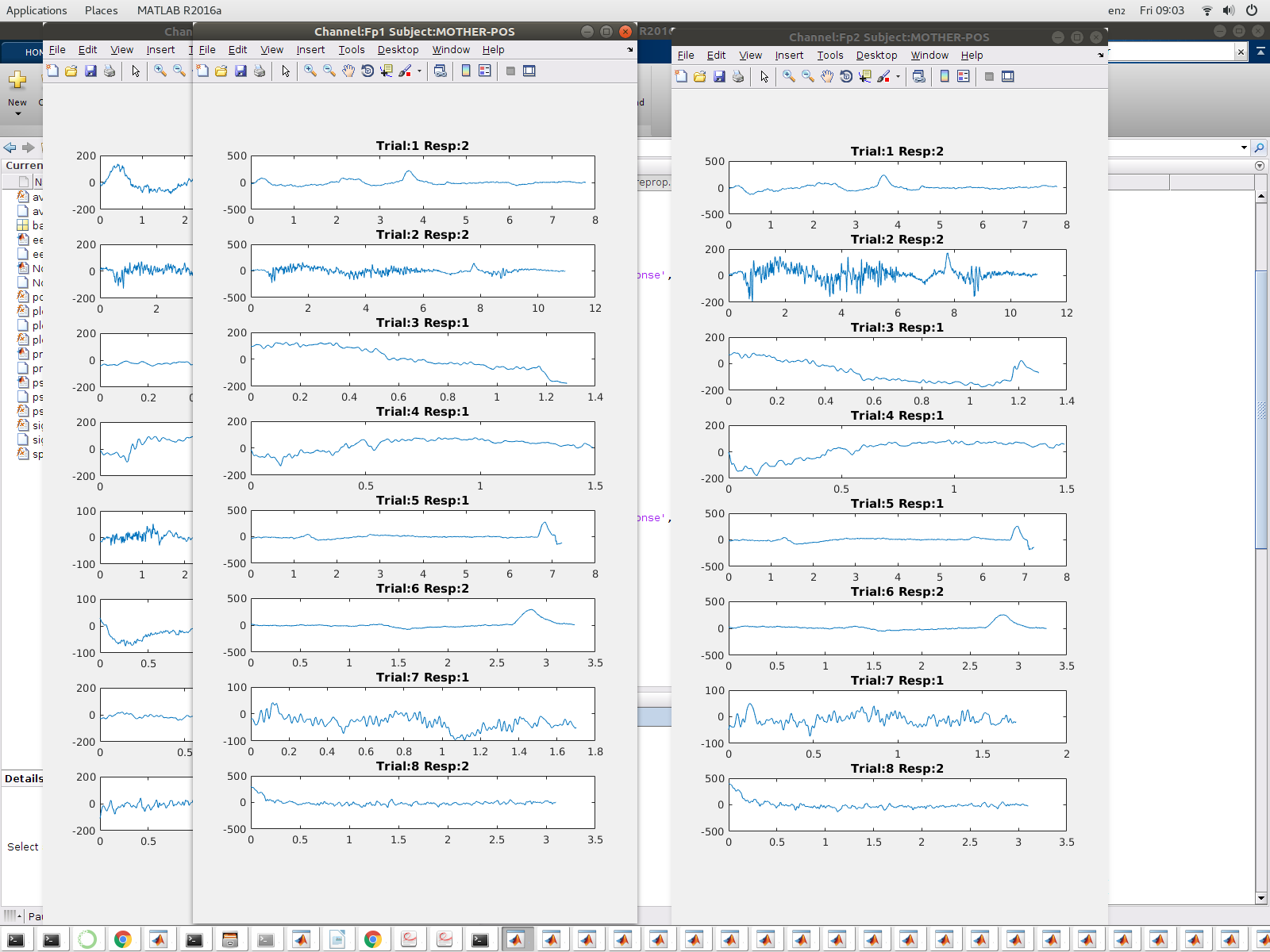
This module is contained in the script eeglx.m. Excecuting this script will open the file and extract each trial signals allocating it in arrays which are eventually then processed and analysed. I used the sampling frequency and the length of the signal to construct the respecitve time domains, as show in the image below.
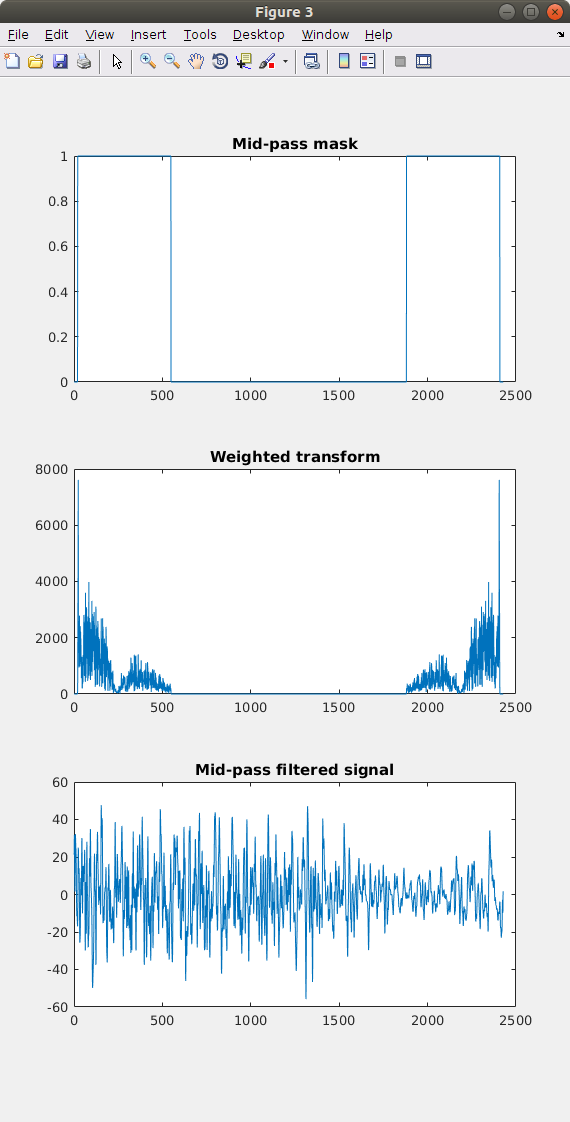
Once the data has ben loaded and stored in matrices in the workspace the script preprop.m detrends and returns spectra for each channnel and arrays of regularised signals. This script has to be exceuted per EEG set.
The final step is to excecute avp.m which averages. Then psel.m evaluates the spectral estimator and classifier.
The averages, estimator and classifier codes are contained in pclass.m, logclass.m
All the other files are for plotting.