dockstore-cgpwgs provides a complete multi threaded WGS analysis for SNV, INDEL, SV and Copynumber
variation with associated annotation of VCF files. This has been packaged specifically for use with
the Dockstore.org framework.
| Master | Develop |
|---|---|
- Usage
- Other uses
- Verifying your deployment
- Example data
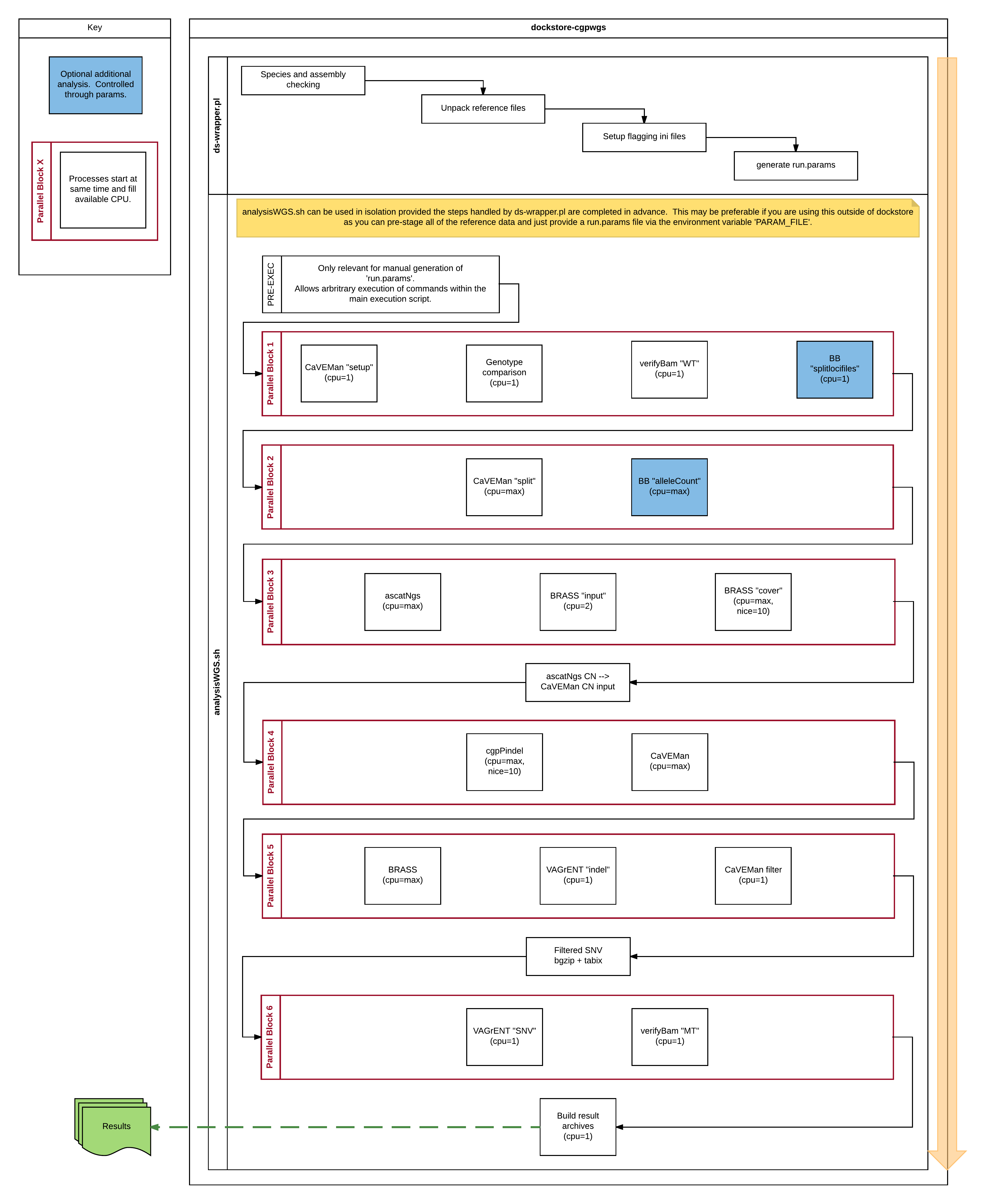
- Diagram of internals
- Development environment
- Release process
- LICENCE
This is intended to be run using the Dockstore.org framework under docker but can be executed as a normal docker container (or imported into singularity.
See the dockstore execution method here.
You should see the usage for the ds-cgpwgs.pl script for all parameters (or the cwl definition).
Required input files are
- Tumour BAM file
- Normal BAM file
- Core reference archive (e.g. core_ref_GRCh37d5.tar.gz)
- WXS reference archive (e.g. SNV_INDEL_ref_GRCh37d5.tar.gz)
- WGS reference archive (e.g. CNV_SV_ref_GRCh37d5_brass6+.tar.gz)
- VAGrENT (annotation) reference archive (e.g. VAGrENT_ref_GRCh37d5_ensembl_75.tar.gz)
- Subclonal reference archive (SUBCL_ref_GRCh37d5.tar.gz)
- Only needed if
skipbbisfalse
- QC reference archive (e.g. qcGenotype_GRCh37d5.tar.gz)
Inputs 1&2 are expected to have been mapped using dockstore-cgpmap
Please check the Wiki then raise an issue if you require additional information on how to generate your own reference files. Much of this information is available on the individual algorithm wiki pages (or the subsequently linked protocols papers).
- Specific to dockstore-cgpwgs:
- Inherited from dockstore-cgpwxs:
When running outside of a docker container you can set the number of CPUs via:
export CPU=N-cores|-coption ofds-cgpwgs.pl
If not set detects available cores on system.
All of the tools installed as part of dockstore-cgpmap and dockstore-cgpwxs and the above packages are available for direct use.
See the docker guide in the wiki for more details.
The resulting docker container has been tested with Singularity.
See the docker guide in the wiki for more details.
The examples/ tree contains test json files populated with data that can be used to verify the
tool.
The data linked in the 'examples' area is from the cell line COLO-829.
This diagram was generated based on v1.1.0, it does not describe any of the file provisioning handled by a Dockstore run.
This project uses git pre-commit hooks. Please enable them to prevent inappropriate large files being included. Any pull request found not to have adhered to this will be rejected and the branch will need to be manually cleaned to keep the repo size down.
Activate the hooks with
git config core.hooksPath git-hooksThis project is maintained using HubFlow.
- Make appropriate changes
- Build image locally
- Run all example inputs and verify any changes are acceptable
- Bump version in
DockerfileandDockstore.cwl - Push changes
- Check state on Travis
- Generate the release (add notes to GitHub)
- Confirm that image has been built on quay.io
- Update the dockstore entry, see their docs.
Copyright (c) 2017-2019 Genome Research Ltd.
Author: Cancer Genome Project <cgphelp@sanger.ac.uk>
This file is part of dockstore-cgpwgs.
dockstore-cgpwgs is free software: you can redistribute it and/or modify it under
the terms of the GNU Affero General Public License as published by the Free
Software Foundation; either version 3 of the License, or (at your option) any
later version.
This program is distributed in the hope that it will be useful, but WITHOUT
ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS
FOR A PARTICULAR PURPOSE. See the GNU Affero General Public License for more
details.
You should have received a copy of the GNU Affero General Public License
along with this program. If not, see <http://www.gnu.org/licenses/>.
1. The usage of a range of years within a copyright statement contained within
this distribution should be interpreted as being equivalent to a list of years
including the first and last year specified and all consecutive years between
them. For example, a copyright statement that reads ‘Copyright (c) 2005, 2007-
2009, 2011-2012’ should be interpreted as being identical to a statement that
reads ‘Copyright (c) 2005, 2007, 2008, 2009, 2011, 2012’ and a copyright
statement that reads ‘Copyright (c) 2005-2012’ should be interpreted as being
identical to a statement that reads ‘Copyright (c) 2005, 2006, 2007, 2008,
2009, 2010, 2011, 2012’."