Scripts and modules for training and testing deep neural networks for ECG automatic classification. Companion code to the paper "Screening for Chagas disease from the electrocardiogram using a deep neural network".
Citation: TBA
Four different cohorts are used in the study. More detailed information TBP.
- The
CODEstudy cohort is used for training. - The
SaMi-Tropcohort is used for training. - The
ELSA-Brasilcohort is used for testing. - The
REDS-IIcohort is used for testing.
The code training and evaluation is implemented in Python.
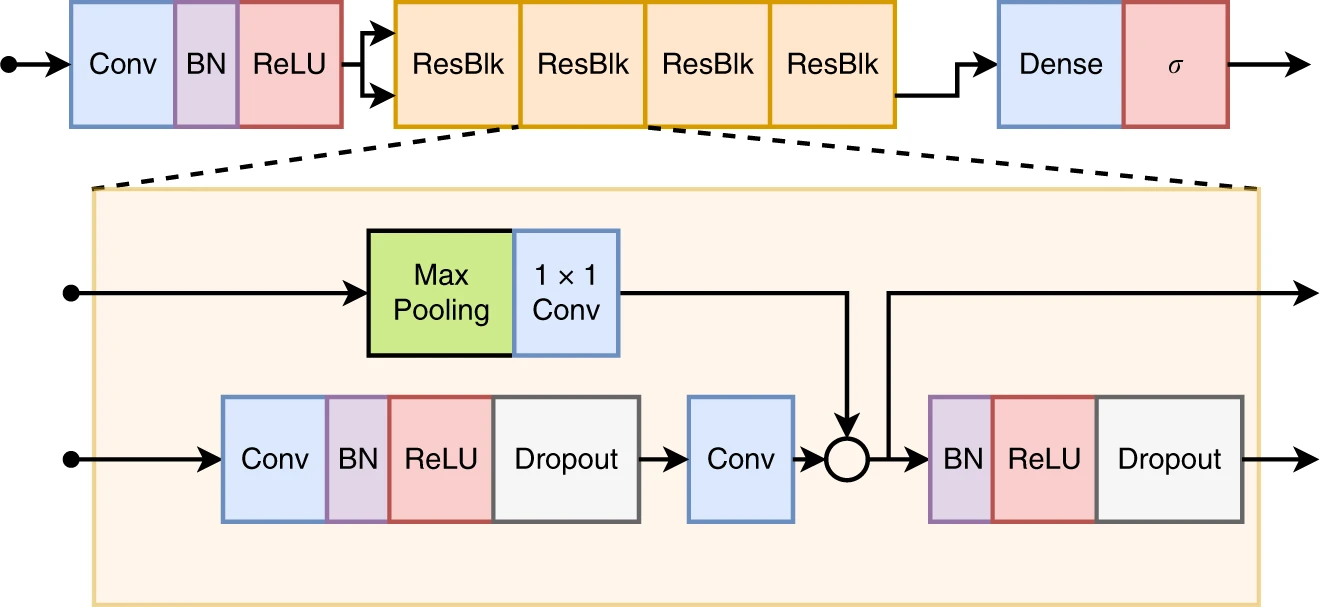
The model used in the paper is a residual neural network. The architecture implementation
in pytorch is available in resnet.py. It follows closely
this architecture.
The model can be trained using the script train.py. Alternatively, pre-trained weighs as described in the paper is available at https://doi.org/10.5281/zenodo.7371623.
- model input:
shape = (N, 12, 4096). The input tensor should contain the 4096 points of the ECG tracings sampled at 400Hz (i.e., a signal of approximately 10 seconds). Both in the training and in the test set, when the signal was not long enough, we filled the signal with zeros, so 4096 points were attained. The last dimension of the tensor contains points of the 12 different leads. The leads are ordered in the following order: {DI, DII, DIII, AVR, AVL, AVF, V1, V2, V3, V4, V5, V6}. All signal are represented as 32 bits floating point numbers at the scale 1e-4V: so if the signal is in V it should be multiplied by 1000 before feeding it to the neural network model. - model output:
shape = (N, 1). With the entry being interpreted as the predicted probability of Chagas disease.
This code was tested on Python 3 with Pytorch 1.9. It uses numpy, pandas,
h5py for loading and processing the data and matplotlib and seaborn
for the plots. See requirements.txt to see a full list of requirements
and library versions.
-
train.py: Script for training the neural network. To train the neural network run. -
evaluate.py: Script for generating the neural network predictions. -
ensemble_merge.py: Script for merging the results of multiple models. -
evaluate_from_file.py: Script for computing metrics and CIs. -
generate_visualisations.py: Script for generating heat maps through grad-cam analysis. -
generate_figs.py: Script for generating result figures (ROC, precision recall, histograms, training). -
stratification.py: Script for generating stratified results wrt age and sex. -
run_code.py: Script demonstrating how to train, evaluate and compute results from the command line. -
resnet.py: Auxiliary module that defines the architecture of the deep neural network.