This is a project repository for our paper-
- Subedi, S. and Park, Y.P., Single-cell Pairwise Relationships Untangled by Composite Embedding model, iScience, 2023.
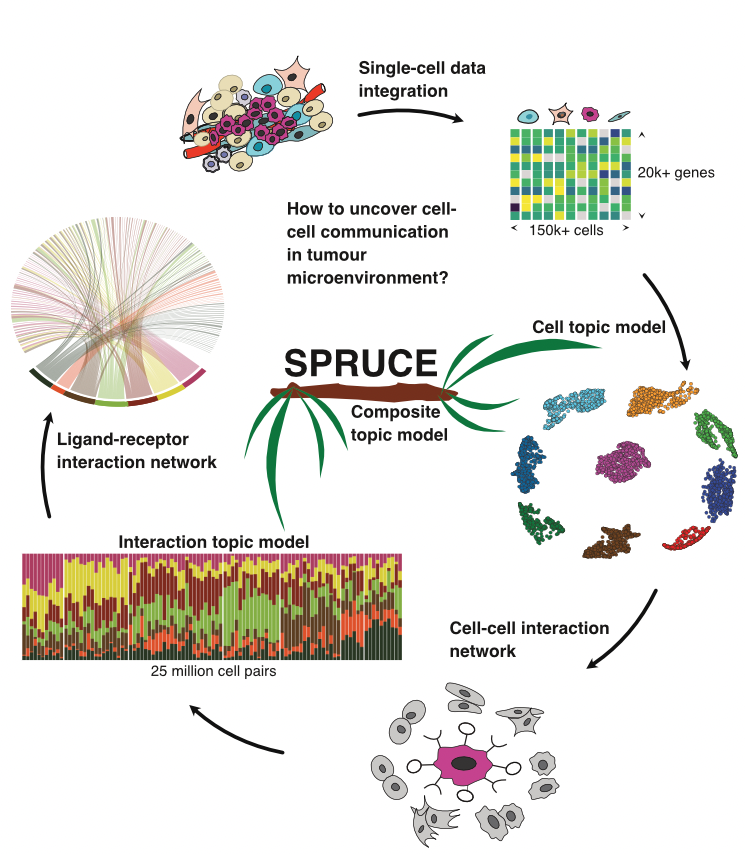
In multi-cellular organisms, cell identity and functions are primed and refined through interactions with other surrounding cells. Here, we propose a scalable machine learning method, termed SPURCE, which is designed to systematically ascertain common cell-cell communication patterns embedded in single-cell RNA-seq data. We applied our approach to investigate tumour microenvironments consolidating multiple breast cancer data sets and found seven frequently-observed interaction signatures and underlying gene-gene interaction networks. Our results implicate that a part of tumour heterogeneity, especially within the same subtype, is better understood by differential interaction patterns rather than the static expression of known marker genes.
- python - numpy, pandas, scipy, sklearn, annoy, pytorch, igraph, seaborn
- R - celldex, SingleR, SingleCellExperiment, ggplot2, pheatmap, circlize, bipartite
- Clone the repo
git clone https://github.com/causalpathlab/spruceTopic.git