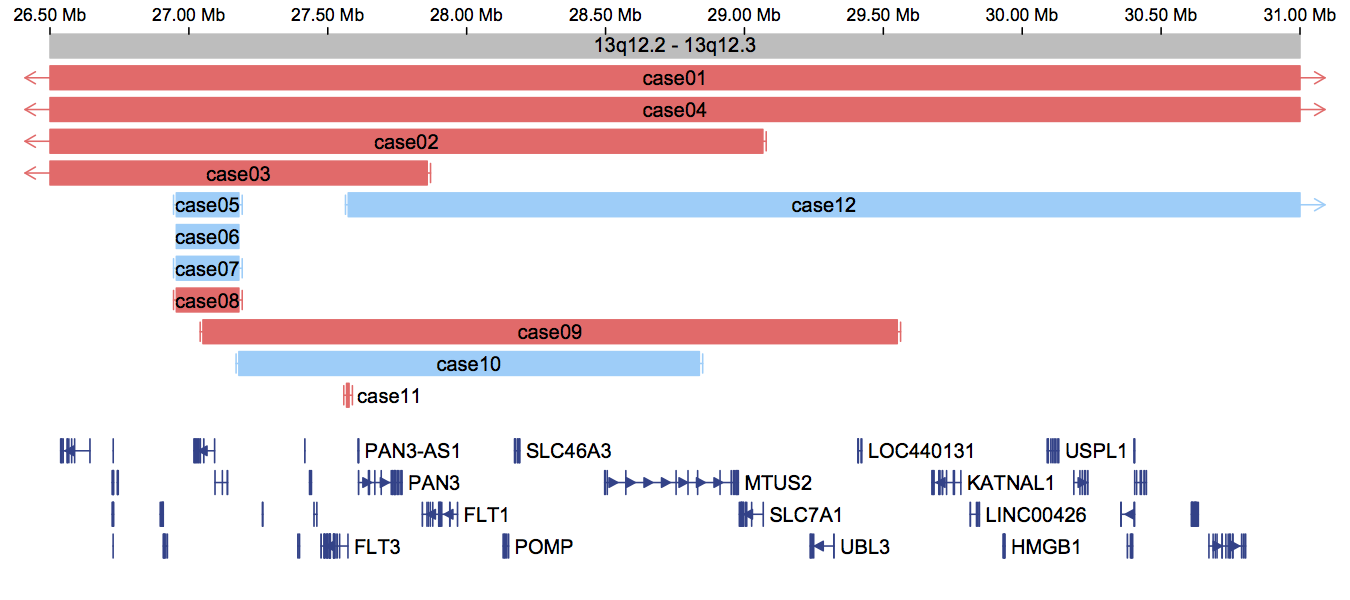
Draw CNV diagrams
Program: cnvgram
Version: 0.0.1
Author: Colby Chiang (cc2qe@virginia.edu)
Date: 11/29/2011
Affiliation: UVA, CHGR, MGH
Description: cnvgram is a script written in R to conveniently draw chromosomal ideograms with copy number variants and genes.
To see an example, navigate to the directory example and open cnvgram_example.R (the driver script). You must point the source (first line of the cnvgram_example.R file) to the correct path of cnvgram.R (the source file), otherwise it won't run.
The function "importCnvs" in the driver file references a 16 column text file with data for all the CNVs you want to draw.
Col Description
1 ID that will be printed on the graph
2 Reference ID that will not be shown on the graph
3 textAlign can be "left", "right", "center", or "none" to indicate where the CNV should be labeled
4 Event must be either "Copy Gain" or "Copy Loss" or "Special". "Special" is deprecated, but will probably make your CNV blue.
5 Phenotype (ignored by CNV Draw)
6 Chrom band (ignored by CNV Draw)
7 chromosome
8 CNV start coord
9 CNV end coord
10 Size of CNV (ignored by CNV Draw)
11 Left error bar (-1 for no error bar)
12 Right error bar (-1 for no error bar)
13 Row to draw the CNV
14 number of gains (deprecated, used to be used by "Special")
15 number of losses (deprecated, used to be used by "Special")
16 1 or 0. CNV Draw will draw diagonal shading for gene specific events.
The function "importGenes" in the driver file references an 18 column text file with data for the genes in the region and their metadata.
Col Description
1-16 The columns of refSeq genes as downloaded from the UCSC genome browser
17 Row to draw the gene
18 position of label: "left", "right", "center", or "none"
To get the first 16 columns, you can download the refGene data from UCSC at the following location: http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/refGene.txt.gz
It's often useful to filter this down to just get genes in your region. I use this quick awk script
curl -s http://hgdownload.cse.ucsc.edu/goldenpath/hg19/database/refGene.txt.gz | gzip -cdfq | awk 'BEGIN {FS="\t"; OFS="\t"} { if ($3=="chr2" && $5>=144750000 && $6<=152400000) print }' | sort -nk5,5 | awk 'BEGIN {FS="\t"; OFS="\t"; GENE="INITIATE"; MAX_SIZE=0; STORED_ROW=""} { if ($13==GENE && ($6-$5)>MAX_SIZE) {MAX_SIZE=($6-$5); STORED_ROW=$0} else { if (GENE!=$13) { if (GENE!="INITIATE") {print STORED_ROW;} GENE=$13; STORED_ROW=$0 } } } END { print STORED_ROW}'
Don't forget to add columns 17 and 18 before trying to load it into importGenes though.