To be able to answer where the genetic variants on genome, or how many there are and whether one could classify them, I cover the following techniques:
-
To find SNPs and indels in sequencing data using VariantTools
-
To predict ORF in long reference sequences
-
To plot features on genetic maps with karyoploteR
-
To find alternative transcript isoforms
-
To select and classify variants using VariantAnnotation
-
To extract information in genomic regions of interest
-
To find phenotype and genotype associations with GWAS
-
To estimate the copy number at a locus of interest
For these, I will need the following packages:
-
Biostrings, GenomicRanges, gmapR, karyoploteR, rtracklayer, systemPipeR, SummarizedExperiment, VariantAnnotation, VariantTools from Bioconductor
-
rrBLUP
Here, I will create a pipeline from reads to lists of genes with variants.
# load packages
library(GenomicRanges)
library(gmapR)
library(rtracklayer)
library(VariantAnnotation)
library(VariantTools)
# load the datasets
bam_folder <- file.path(getwd())
bam_folder_contents <- list.files(file.path(getwd()))
bam <- file.path(bam_folder, "hg17_snps.bam")
fasta_file <- file.path(bam_folder,"chr17.83k.fa")
# set up the genome object
fa <- rtracklayer::FastaFile(fasta_file)
genome <- gmapR::GmapGenome(fa, create=TRUE)
# set up the parameter objects
var_params <- VariantCallingFilters(read.count = 19,
p.lower = 0.01)
qual_params <- TallyVariantsParam(genome = genome,
minimum_mapq = 20)
# call the variants
called_variants <- callVariants(bam, qual_params,
calling.filters = var_params)
head(called_variants)
## VRanges object with 6 ranges and 17 metadata columns:
## seqnames ranges strand ref alt totalDepth
## <Rle> <IRanges> <Rle> <character> <characterOrRle> <integerOrRle>
## [1] NC_000017.10 64 * G T 759
## [2] NC_000017.10 69 * G T 812
## [3] NC_000017.10 70 * G T 818
## [4] NC_000017.10 73 * T A 814
## [5] NC_000017.10 77 * T A 802
## [6] NC_000017.10 78 * G T 798
## refDepth altDepth sampleNames softFilterMatrix | n.read.pos
## <integerOrRle> <integerOrRle> <factorOrRle> <matrix> | <integer>
## [1] 739 20 <NA> | 17
## [2] 790 22 <NA> | 19
## [3] 796 22 <NA> | 20
## [4] 795 19 <NA> | 13
## [5] 780 22 <NA> | 19
## [6] 777 21 <NA> | 17
## n.read.pos.ref raw.count.total count.plus count.plus.ref count.minus
## <integer> <integer> <integer> <integer> <integer>
## [1] 64 759 20 739 0
## [2] 69 812 22 790 0
## [3] 70 818 22 796 0
## [4] 70 814 19 795 0
## [5] 70 802 22 780 0
## [6] 70 798 21 777 0
## count.minus.ref count.del.plus count.del.minus read.pos.mean
## <integer> <integer> <integer> <numeric>
## [1] 0 0 0 30.9000
## [2] 0 0 0 40.7273
## [3] 0 0 0 34.7727
## [4] 0 0 0 36.1579
## [5] 0 0 0 38.3636
## [6] 0 0 0 39.7143
## read.pos.mean.ref read.pos.var read.pos.var.ref mdfne mdfne.ref
## <numeric> <numeric> <numeric> <numeric> <numeric>
## [1] 32.8755 318.558 347.804 NA NA
## [2] 35.4190 377.004 398.876 NA NA
## [3] 36.3442 497.762 402.360 NA NA
## [4] 36.2176 519.551 402.843 NA NA
## [5] 36.0064 472.327 397.070 NA NA
## [6] 35.9241 609.076 390.463 NA NA
## count.high.nm count.high.nm.ref
## <integer> <integer>
## [1] 20 738
## [2] 22 789
## [3] 22 796
## [4] 19 769
## [5] 22 780
## [6] 21 777
## -------
## seqinfo: 1 sequence from chr17.83k genome
## hardFilters(4): nonRef nonNRef readCount likelihoodRatio
# functions to read annotation from files
get_annotated_regions <- function(file_name){
gff <- import.gff(file_name)
as(gff, "GRanges")
}
get_annotated_regions2 <- function(file_name){
bed <- import.bed(file_name)
as(bed, "GRanges")
}
# get annotations
genes <- get_annotated_regions("chr17.83k.gff3")
# calculate overlapping variants
overlaps <- findOverlaps(called_variants, genes)
overlaps
## Hits object with 12684 hits and 0 metadata columns:
## queryHits subjectHits
## <integer> <integer>
## [1] 35176 1
## [2] 35176 2
## [3] 35176 3
## [4] 35177 1
## [5] 35177 2
## ... ... ...
## [12680] 40944 2
## [12681] 40944 7
## [12682] 40945 1
## [12683] 40945 2
## [12684] 40945 7
## -------
## queryLength: 44949 / subjectLength: 8
# as a list
genes[subjectHits(overlaps)]
## GRanges object with 12684 ranges and 20 metadata columns:
## seqnames ranges strand | source type score
## <Rle> <IRanges> <Rle> | <factor> <factor> <numeric>
## [1] NC_000017.10 64099-76866 - | havana ncRNA_gene NA
## [2] NC_000017.10 64099-76866 - | havana lnc_RNA NA
## [3] NC_000017.10 64099-65736 - | havana exon NA
## [4] NC_000017.10 64099-76866 - | havana ncRNA_gene NA
## [5] NC_000017.10 64099-76866 - | havana lnc_RNA NA
## ... ... ... ... . ... ... ...
## [12680] NC_000017.10 64099-76866 - | havana lnc_RNA NA
## [12681] NC_000017.10 76723-76866 - | havana exon NA
## [12682] NC_000017.10 64099-76866 - | havana ncRNA_gene NA
## [12683] NC_000017.10 64099-76866 - | havana lnc_RNA NA
## [12684] NC_000017.10 76723-76866 - | havana exon NA
## phase ID Name biotype
## <integer> <character> <character> <character>
## [1] <NA> gene:ENSG00000280279 AC240565.2 lincRNA
## [2] <NA> transcript:ENST00000623180 AC240565.2-201 lincRNA
## [3] <NA> <NA> ENSE00003759547 <NA>
## [4] <NA> gene:ENSG00000280279 AC240565.2 lincRNA
## [5] <NA> transcript:ENST00000623180 AC240565.2-201 lincRNA
## ... ... ... ... ...
## [12680] <NA> transcript:ENST00000623180 AC240565.2-201 lincRNA
## [12681] <NA> <NA> ENSE00003756684 <NA>
## [12682] <NA> gene:ENSG00000280279 AC240565.2 lincRNA
## [12683] <NA> transcript:ENST00000623180 AC240565.2-201 lincRNA
## [12684] <NA> <NA> ENSE00003756684 <NA>
## description gene_id logic_name version
## <character> <character> <character> <character>
## [1] novel transcript ENSG00000280279 havana 1
## [2] <NA> <NA> <NA> 1
## [3] <NA> <NA> <NA> 1
## [4] novel transcript ENSG00000280279 havana 1
## [5] <NA> <NA> <NA> 1
## ... ... ... ... ...
## [12680] <NA> <NA> <NA> 1
## [12681] <NA> <NA> <NA> 1
## [12682] novel transcript ENSG00000280279 havana 1
## [12683] <NA> <NA> <NA> 1
## [12684] <NA> <NA> <NA> 1
## Parent tag transcript_id
## <CharacterList> <character> <character>
## [1] <NA> <NA>
## [2] gene:ENSG00000280279 basic ENST00000623180
## [3] transcript:ENST00000623180 <NA> <NA>
## [4] <NA> <NA>
## [5] gene:ENSG00000280279 basic ENST00000623180
## ... ... ... ...
## [12680] gene:ENSG00000280279 basic ENST00000623180
## [12681] transcript:ENST00000623180 <NA> <NA>
## [12682] <NA> <NA>
## [12683] gene:ENSG00000280279 basic ENST00000623180
## [12684] transcript:ENST00000623180 <NA> <NA>
## transcript_support_level constitutive ensembl_end_phase ensembl_phase
## <character> <character> <character> <character>
## [1] <NA> <NA> <NA> <NA>
## [2] 5 <NA> <NA> <NA>
## [3] <NA> 1 -1 -1
## [4] <NA> <NA> <NA> <NA>
## [5] 5 <NA> <NA> <NA>
## ... ... ... ... ...
## [12680] 5 <NA> <NA> <NA>
## [12681] <NA> 1 -1 -1
## [12682] <NA> <NA> <NA> <NA>
## [12683] 5 <NA> <NA> <NA>
## [12684] <NA> 1 -1 -1
## exon_id rank
## <character> <character>
## [1] <NA> <NA>
## [2] <NA> <NA>
## [3] ENSE00003759547 5
## [4] <NA> <NA>
## [5] <NA> <NA>
## ... ... ...
## [12680] <NA> <NA>
## [12681] ENSE00003756684 1
## [12682] <NA> <NA>
## [12683] <NA> <NA>
## [12684] ENSE00003756684 1
## -------
## seqinfo: 1 sequence from an unspecified genome; no seqlengths
Here, I will be predicting open reading frames using systemPipeR.
# load the libraries
library(Biostrings)
library(systemPipeR)
# input genome
dna_object <- readDNAStringSet("arabidopsis_chloroplast.fa")
# predict Open Reading Frames (ORF)
predicted_orfs <- predORF(dna_object,
n = "all",
type = "gr",
mode = "ORF",
strand = "both",
longest_disjoint = TRUE)
predicted_orfs
## GRanges object with 2501 ranges and 2 metadata columns:
## seqnames ranges strand | subject_id inframe2end
## <Rle> <IRanges> <Rle> | <integer> <numeric>
## 1 chloroplast 86762-93358 + | 1 2
## 1162 chloroplast 2056-2532 - | 1 3
## 2 chloroplast 72371-73897 + | 2 2
## 1163 chloroplast 77901-78362 - | 2 1
## 3 chloroplast 54937-56397 + | 3 3
## ... ... ... ... . ... ...
## 2497 chloroplast 129757-129762 - | 1336 3
## 2498 chloroplast 139258-139263 - | 1337 3
## 2499 chloroplast 140026-140031 - | 1338 3
## 2500 chloroplast 143947-143952 - | 1339 3
## 2501 chloroplast 153619-153624 - | 1340 3
## -------
## seqinfo: 1 sequence from an unspecified genome; no seqlengths
GRanges object returened 2501 ORFs, thus, next step is to filter out some ORFs that occured by chance from the sequence.
# calculate the properties of the reference genome
bases <- c("A", "C", "G", "T")
raw_seq_string <- strsplit(as.character(dna_object), "")
seqlength <- width(dna_object[1])
counts <- lapply(bases, function(x){sum(grepl(x, raw_seq_string))})
probs <- unlist(lapply(counts, function(base_count){signif(base_count / seqlength, 2)}))
# find the longest ORF in a simulated genome
get_longest_orf <- function(x,
length = 1000,
probs = c(0.25, 0.25, 0.25, 0.25),
bases = c("A", "C", "G", "T")) {
random_genome <- paste0(sample(bases,
size = length,
replace = TRUE,
prob = probs), collapse = "")
random_dna_object <- DNAStringSet(random_genome)
names(random_dna_object) <- c("random_dna_string")
orfs <- predORF(random_dna_object,
n = 1,
type = "gr",
mode = "ORF",
strand = "both",
longest_disjoint = TRUE)
return(max(width(orfs)))
}
# run the function on 10 simulated genomes
random_lenghts <- unlist(lapply(1:10,
get_longest_orf,
length = seqlength,
probs = probs,
bases = bases))
# find the longest random ORF
longest_random_orf <- max(random_lenghts)
# keep only predicted ORFs longer than the longest random ORF
keep <- width(predicted_orfs) > longest_random_orf
orfs_to_keep <- predicted_orfs[keep]
head(orfs_to_keep)
## GRanges object with 6 ranges and 2 metadata columns:
## seqnames ranges strand | subject_id inframe2end
## <Rle> <IRanges> <Rle> | <integer> <numeric>
## 1 chloroplast 86762-93358 + | 1 2
## 2 chloroplast 72371-73897 + | 2 2
## 3 chloroplast 54937-56397 + | 3 3
## 4 chloroplast 57147-58541 + | 4 1
## 5 chloroplast 33918-35141 + | 5 1
## 6 chloroplast 32693-33772 + | 6 2
## -------
## seqinfo: 1 sequence from an unspecified genome; no seqlengths
# save the ORFs
extracted_orfs <- getSeq(dna_object, orfs_to_keep)
names(extracted_orfs) <- paste0("orf_", 1:length(orfs_to_keep))
writeXStringSet(extracted_orfs, "saved_orfs.fa")
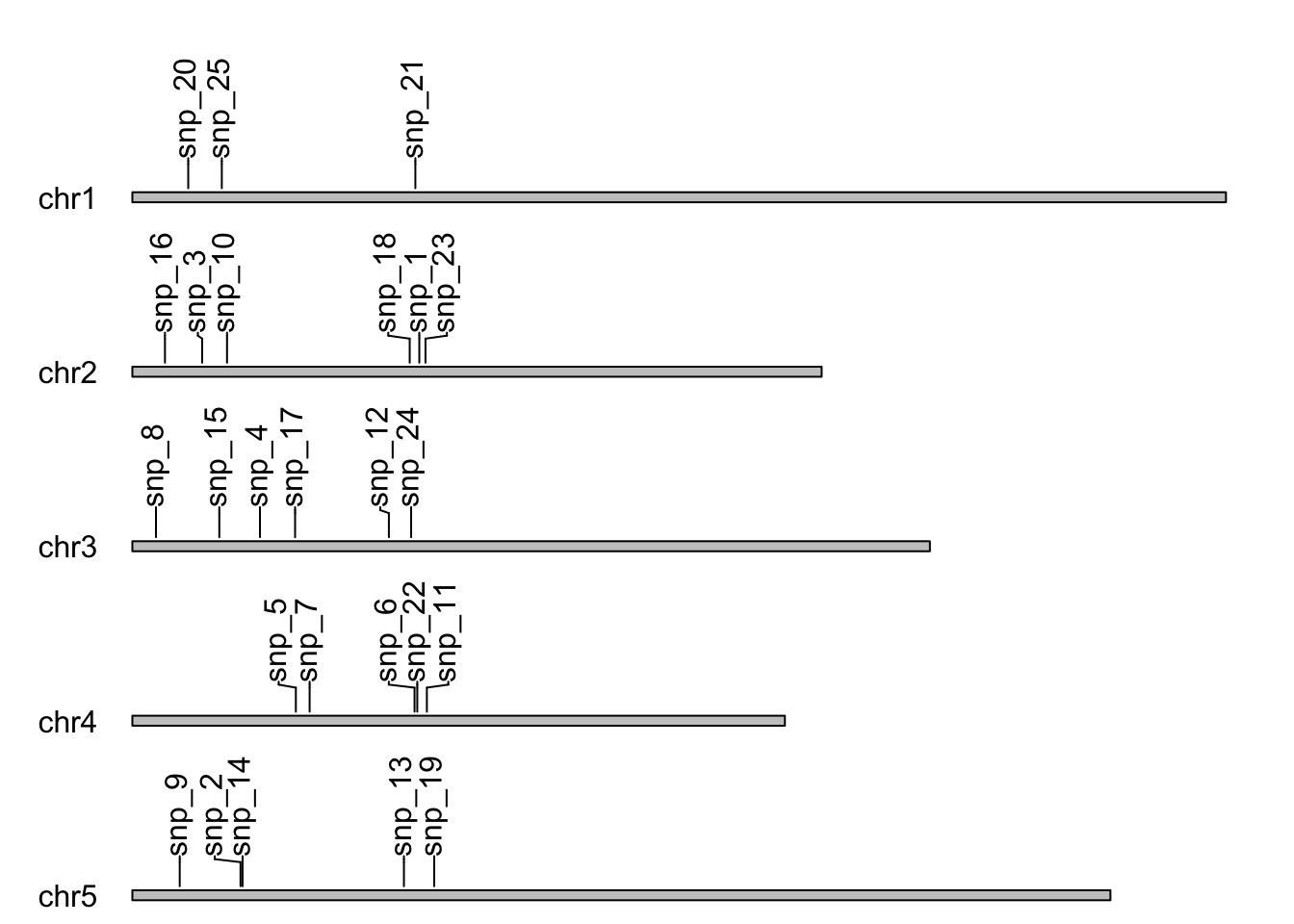
Here, I will create some visualisations for chromosomes and ideograms using karyoploteR package.
# load packages
library(karyoploteR)
library(GenomicRanges)
# set up the genome object
genome_df <- data.frame(
chr = paste0("chr", 1:5),
start = rep(1, 5),
end = c(34964571, 22037565, 25499034, 20862711, 31270811)
)
genome_gr <- makeGRangesFromDataFrame(genome_df)
# set up the SNP positions
snp_pos <- sample(1:1e7, 25)
snps <- data.frame(
chr = paste0("chr", sample(1:5, 25, replace = TRUE)),
start = snp_pos,
end = snp_pos
)
snps_gr <- makeGRangesFromDataFrame(snps)
# create labels
snp_labels <- paste0("snp_", 1:25)
# set the plot margins
plot.params <- getDefaultPlotParams(plot.type = 1)
plot.params$data1outmargin <- 600
# create the base plot and add tracks
kp <- plotKaryotype(genome = genome_gr,
plot.type = 1,
plot.params = plot.params)
kpPlotMarkers(kp, snps_gr, labels = snp_labels)
Using a VCF file, I will do subsequent analyses that need further filtering and classification based on features of the individual variants like the depth coverage in the alternative allele.
# load packages
library(VariantAnnotation)
# create a prefilter function
is_not_microsat <- function(x){
!grepl("microsat", x, fixed = TRUE)
}
# load up the prefilter function into a FilterRules object
prefilters <- FilterRules(list(microsat = is_not_microsat))
# create a filter function to keep variants where the reference allele is in less than half the reads
major_alt <- function(x){
af <- info(x)$AF
result <- unlist(lapply(af, function(x){
x[1] < 0.5
}))
return(result)
}
# load the filter function into a FilterRules object
filters <- FilterRules(list(alt_is_major = major_alt))
# load the input VCF file and apply filters
vcf_file <- "sample.vcf.gz"
filterVcf(vcf_file,
"hg17",
"filtered.vcf",
prefilters = prefilters,
filters = filters)
Here, I will look in more detail at data that falls in a particular genomic region of interest, whether that be the SNPs and variants in a gene or the genes in a particular locus.
# load packages
library(GenomicRanges)
library(rtracklayer)
library(SummarizedExperiment)
library(readr)
# define functions that read and create GRanges
# from .GFF
get_granges_gff <- function(file_name){
gff <- import.gff(file_name)
as(gff, "GRanges")
}
# from .BED
get_granges_bed <- function(file_name){
bed <- import.bed(file_name)
as(bed, "GRanges")
}
# from .TXT
get_granges_txt <- function(file_name){
df <- read_tsv(file_name, col_names = TRUE)
makeGRangesFromDataFrame(df, keep.extra.columns = TRUE)
}
# create GRanges objects
gr_from_gff <- get_granges_gff("arabidopsis_chr4.gff")
gr_from_txt <- get_granges_txt("arabidopsis_chr4.txt")
# extract a region by filtering on attributes
genes_on_chr4 <- gr_from_gff[gr_from_gff$type == "gene" & seqnames(gr_from_gff) %in% c("Chr4")]
# manually create a region of interest
region_of_interest_gr <- GRanges(
seqnames = c("Chr4"),
IRanges(c(10000), width = c(1000))
)
# Use the region of interest to subset the larger object
overlap_hits <- findOverlaps(region_of_interest_gr,
gr_from_gff)
features_in_region <- gr_from_gff[subjectHits(overlap_hits)]
head(features_in_region)
## GRanges object with 6 ranges and 10 metadata columns:
## seqnames ranges strand | source type score phase
## <Rle> <IRanges> <Rle> | <factor> <factor> <numeric> <integer>
## [1] Chr4 1-18585056 * | TAIR10 chromosome NA <NA>
## [2] Chr4 2895-10455 - | TAIR10 gene NA <NA>
## [3] Chr4 3895-10455 - | TAIR10 mRNA NA <NA>
## [4] Chr4 4107-10364 - | TAIR10 protein NA <NA>
## [5] Chr4 10365-10455 - | TAIR10 five_prime_UTR NA <NA>
## [6] Chr4 10211-10364 - | TAIR10 CDS NA 0
## ID Name Note
## <character> <character> <CharacterList>
## [1] Chr4 Chr4
## [2] AT4G00020 AT4G00020 protein_coding_gene
## [3] AT4G00020.1 AT4G00020.1
## [4] AT4G00020.1-Protein AT4G00020.1
## [5] <NA> <NA>
## [6] <NA> <NA>
## Parent Index Derives_from
## <CharacterList> <character> <character>
## [1] <NA> <NA>
## [2] <NA> <NA>
## [3] AT4G00020 1 <NA>
## [4] <NA> AT4G00020.1
## [5] AT4G00020.1 <NA> <NA>
## [6] AT4G00020.1,AT4G00020.1-Protein <NA> <NA>
## -------
## seqinfo: 1 sequence from an unspecified genome; no seqlengths
To find genetic variants in many samples, I will use genome-wide association studies method.
# load packages
library(VariantAnnotation)
library(rrBLUP)
# get the VCF file
set.seed(1234)
vcf_file <- "small_sample.vcf"
vcf <- readVcf(vcf_file, "hg19")
# extract the genotype, sample and marker position
gts <- geno(vcf)$GT
samples <- samples(header(vcf))
markers <- rownames(gts)
chrom <- as.character(seqnames(rowRanges(vcf)))
pos <- as.numeric(start(rowRanges(vcf)))
# create a custom function to convert VCF genotypes into the convention used by the GWAS function
# 0/0 means homozygous in VCF file which corresponds to 1 in GWAS function
convert <- function(v){
v <- gsub("0/0", 1, v)
v <- gsub("0/1", 0, v)
v <- gsub("1/0", 0, v)
v <- gsub("1/1", -1, v)
return(v)
}
# call the function and convert result into a numeric matrix
gt_char <- apply(gts, convert, MARGIN = 2)
genotype_matrix <- matrix(as.numeric(gt_char),
nrow(gt_char))
colnames(genotype_matrix) <- samples
# build a dataframe describing the variant
variant_info <- data.frame(marker = markers,
chrom = chrom,
pos = pos)
#build a combined variant/genotype dataframe
genotypes <- cbind(variant_info,
as.data.frame(genotype_matrix))
head(genotypes)
## marker chrom pos NA00001 NA00002 NA00003
## 1 rs6054257 20 14370 1 0 -1
## 2 20:17330_T/A 20 17330 1 0 1
## 3 20:1230237_T/G 20 1230237 1 1 0
# build a phenotype dataframe
phenotypes <- data.frame(
line = samples,
score = rnorm(length(samples))
)
head(phenotypes)
# run GWAS
GWAS(phenotypes, genotypes, plot = FALSE)
## marker chrom pos score
## 1 rs6054257 20 14370 0.3010543
## 2 20:17330_T/A 20 17330 0.3010057
## 3 20:1230237_T/G 20 1230237 0.1655498
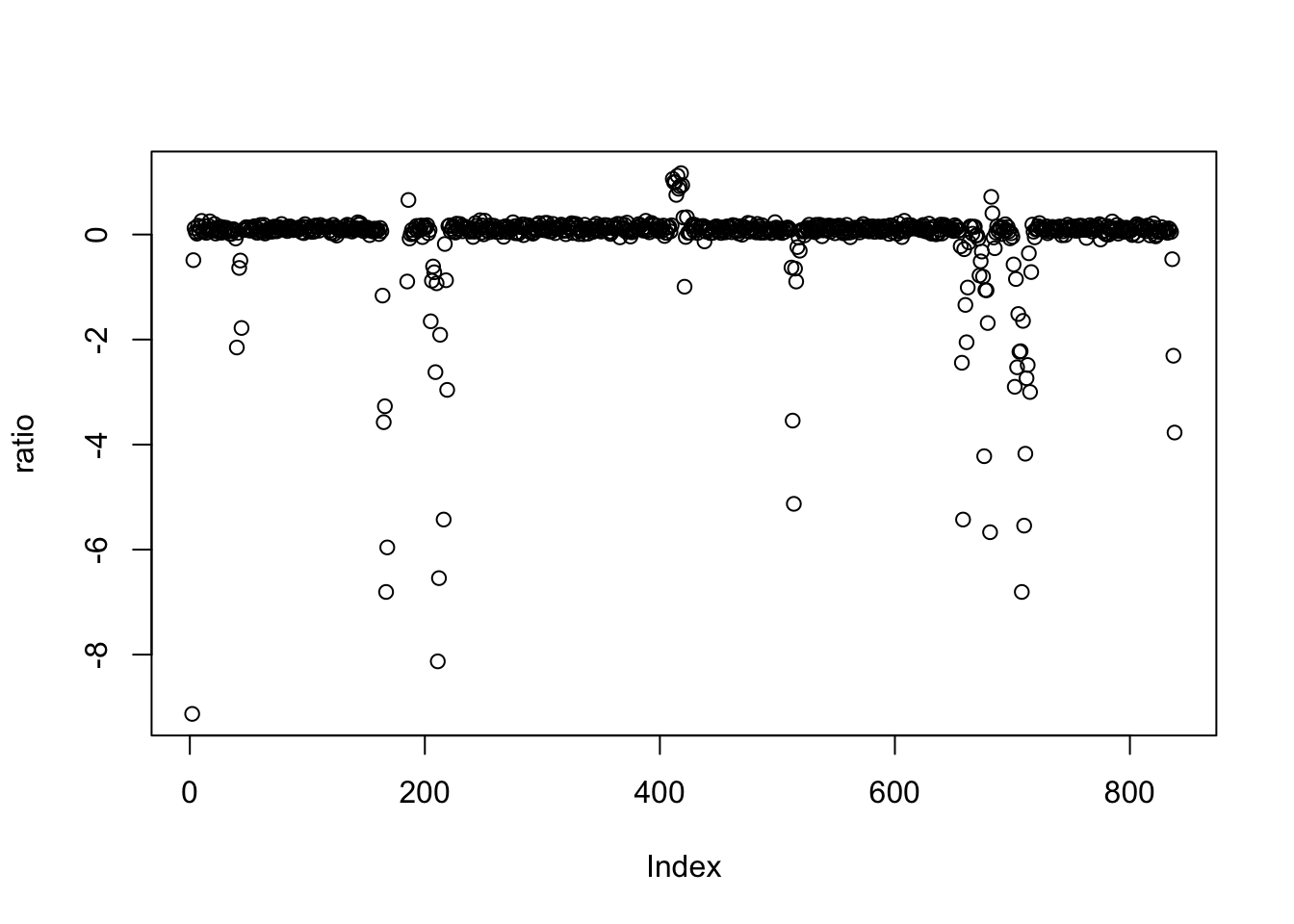
It is often of interest to know how often a sequence occurs in a sample of interest—that is, to estimate whether a locus has been duplicated or its copy number has increased.
# load packages
library(csaw)
library(SummarizedExperiment)
# get counts across the genome
whole_genome <- windowCounts("hg17_snps.bam",
bin = TRUE,
filter = 0,
width = 100,
param = readParam(minq = 20,
dedup = TRUE,
pe = "both"))
colnames(whole_genome) <- c("h17")
# extract data
counts <- assay(whole_genome)[, 1]
# set a low count threshold and set lowers to NA
min_count <- quantile(counts, 0.1)[[1]]
counts[counts <- min_count] <- NA
# double the counts of a set of windows
n <- length(counts)
doubled_windows <- 10
left_pad <- floor((n/2) - doubled_windows)
right_pad <- (n - left_pad - doubled_windows)
multiplier <- c(rep(1, left_pad),
rep(2, doubled_windows),
rep(1, right_pad))
counts <- counts * multiplier
# calculate the mean coverage anad the ratio in each window
mean_cov <- mean(counts, na.rm = TRUE)
ratio <- matrix(log2(counts / mean_cov), ncol = 1)
plot(ratio)
# build summarisedexperiment with new data
se <- SummarizedExperiment(assays = list(ratio),
rowRanges = rowRanges(whole_genome),
colData = c("CoverageRatio"))
# create a region of interest and extract coverage data
region_of_interest <- GRanges(
seqnames = c("NC_000017.10"),
IRanges(c(40700), width = c(1500))
)
overlap_hits <- findOverlaps(region_of_interest, se)
data_in_region <- se[subjectHits(overlap_hits)]
assay(data_in_region)
## [,1]
## [1,] 0.13362884
## [2,] 0.14765840
## [3,] 0.05888607
## [4,] 0.17531474
## [5,] 1.05888607
## [6,] 0.99823847
## [7,] 0.99565531
## [8,] 0.75417705
## [9,] 1.11946151
## [10,] 0.87153400
## [11,] 0.91865791
## [12,] 1.16845020
## [13,] 0.94031227
## [14,] 0.32274511
## [15,] -0.99403968
## [16,] -0.04631696
sessionInfo()
## R version 4.0.1 (2020-06-06)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS Catalina 10.15.6
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_GB.UTF-8/en_GB.UTF-8/en_GB.UTF-8/C/en_GB.UTF-8/en_GB.UTF-8
##
## attached base packages:
## [1] parallel stats4 stats graphics grDevices utils datasets
## [8] methods base
##
## other attached packages:
## [1] csaw_1.22.1 rrBLUP_4.6.1
## [3] readr_1.3.1 karyoploteR_1.14.0
## [5] regioneR_1.20.1 systemPipeR_1.22.0
## [7] ShortRead_1.46.0 GenomicAlignments_1.24.0
## [9] BiocParallel_1.22.0 VariantTools_1.30.0
## [11] VariantAnnotation_1.34.0 SummarizedExperiment_1.18.2
## [13] DelayedArray_0.14.1 matrixStats_0.56.0
## [15] Biobase_2.48.0 rtracklayer_1.48.0
## [17] gmapR_1.30.0 Rsamtools_2.4.0
## [19] Biostrings_2.56.0 XVector_0.28.0
## [21] GenomicRanges_1.40.0 GenomeInfoDb_1.24.2
## [23] IRanges_2.22.2 S4Vectors_0.26.1
## [25] BiocGenerics_0.34.0
##
## loaded via a namespace (and not attached):
## [1] backports_1.1.9 GOstats_2.54.0 Hmisc_4.4-1
## [4] BiocFileCache_1.12.1 lazyeval_0.2.2 GSEABase_1.50.1
## [7] splines_4.0.1 ggplot2_3.3.2 digest_0.6.25
## [10] ensembldb_2.12.1 htmltools_0.5.0 GO.db_3.11.4
## [13] magrittr_1.5 checkmate_2.0.0 memoise_1.1.0
## [16] BSgenome_1.56.0 base64url_1.4 cluster_2.1.0
## [19] limma_3.44.3 annotate_1.66.0 askpass_1.1
## [22] prettyunits_1.1.1 jpeg_0.1-8.1 colorspace_1.4-1
## [25] blob_1.2.1 rappdirs_0.3.1 xfun_0.16
## [28] dplyr_1.0.2 crayon_1.3.4 RCurl_1.98-1.2
## [31] jsonlite_1.7.1 graph_1.66.0 genefilter_1.70.0
## [34] brew_1.0-6 survival_3.2-3 glue_1.4.2
## [37] gtable_0.3.0 zlibbioc_1.34.0 V8_3.2.0
## [40] Rgraphviz_2.32.0 scales_1.1.1 pheatmap_1.0.12
## [43] bezier_1.1.2 DBI_1.1.0 edgeR_3.30.3
## [46] Rcpp_1.0.5 xtable_1.8-4 progress_1.2.2
## [49] htmlTable_2.0.1 foreign_0.8-80 bit_4.0.4
## [52] Formula_1.2-3 rsvg_2.1 AnnotationForge_1.30.1
## [55] htmlwidgets_1.5.1 httr_1.4.2 RColorBrewer_1.1-2
## [58] ellipsis_0.3.1 pkgconfig_2.0.3 XML_3.99-0.5
## [61] nnet_7.3-14 dbplyr_1.4.4 locfit_1.5-9.4
## [64] tidyselect_1.1.0 rlang_0.4.7 AnnotationDbi_1.50.3
## [67] munsell_0.5.0 tools_4.0.1 generics_0.0.2
## [70] RSQLite_2.2.0 evaluate_0.14 stringr_1.4.0
## [73] yaml_2.2.1 knitr_1.29 bit64_4.0.5
## [76] purrr_0.3.4 AnnotationFilter_1.12.0 RBGL_1.64.0
## [79] biomaRt_2.44.1 compiler_4.0.1 rstudioapi_0.11
## [82] curl_4.3 png_0.1-7 tibble_3.0.3
## [85] stringi_1.4.6 GenomicFeatures_1.40.1 lattice_0.20-41
## [88] ProtGenerics_1.20.0 Matrix_1.2-18 vctrs_0.3.4
## [91] pillar_1.4.6 lifecycle_0.2.0 data.table_1.13.0
## [94] bitops_1.0-6 R6_2.4.1 latticeExtra_0.6-29
## [97] hwriter_1.3.2 gridExtra_2.3 dichromat_2.0-0
## [100] assertthat_0.2.1 openssl_1.4.2 Category_2.54.0
## [103] rjson_0.2.20 withr_2.2.0 batchtools_0.9.13
## [106] GenomeInfoDbData_1.2.3 hms_0.5.3 grid_4.0.1
## [109] rpart_4.1-15 bamsignals_1.20.0 DOT_0.1
## [112] rmarkdown_2.3 biovizBase_1.36.0 base64enc_0.1-3