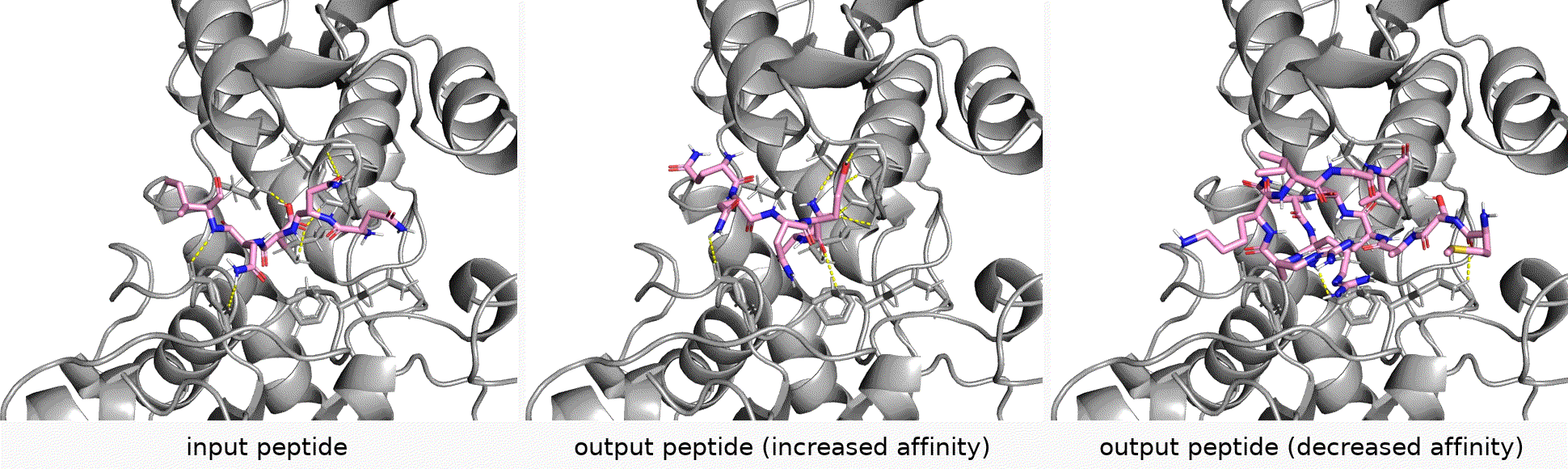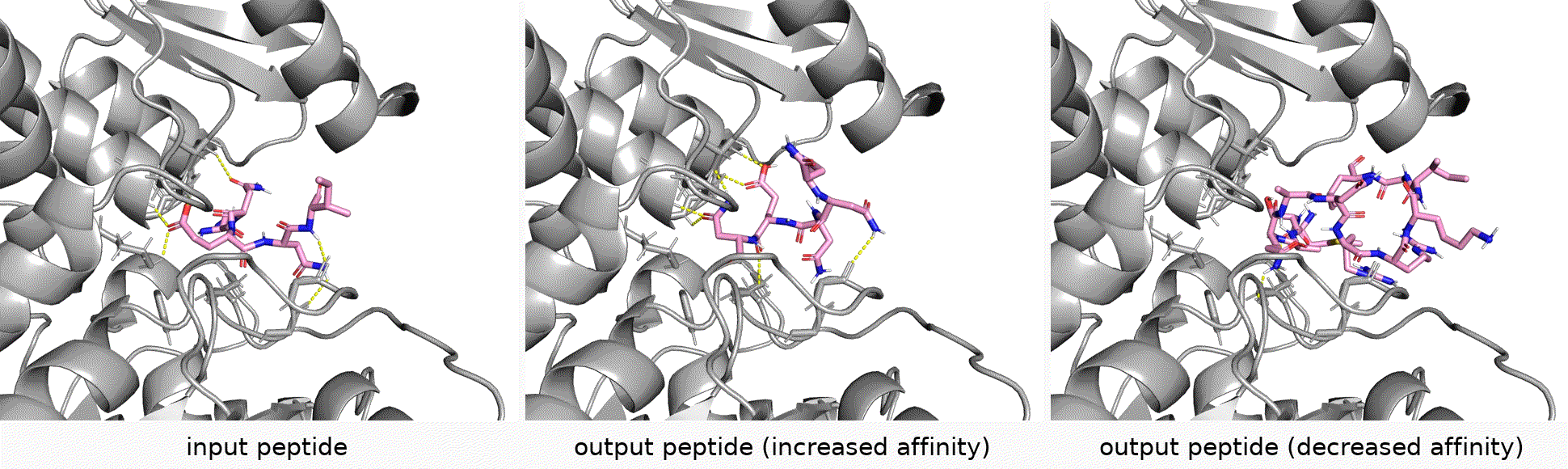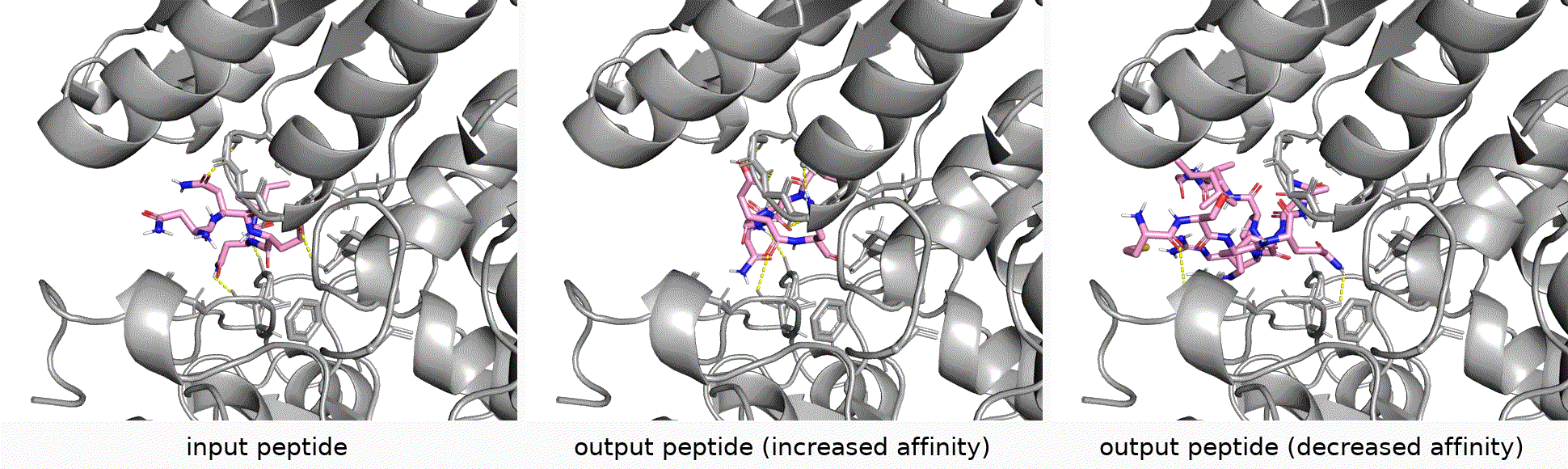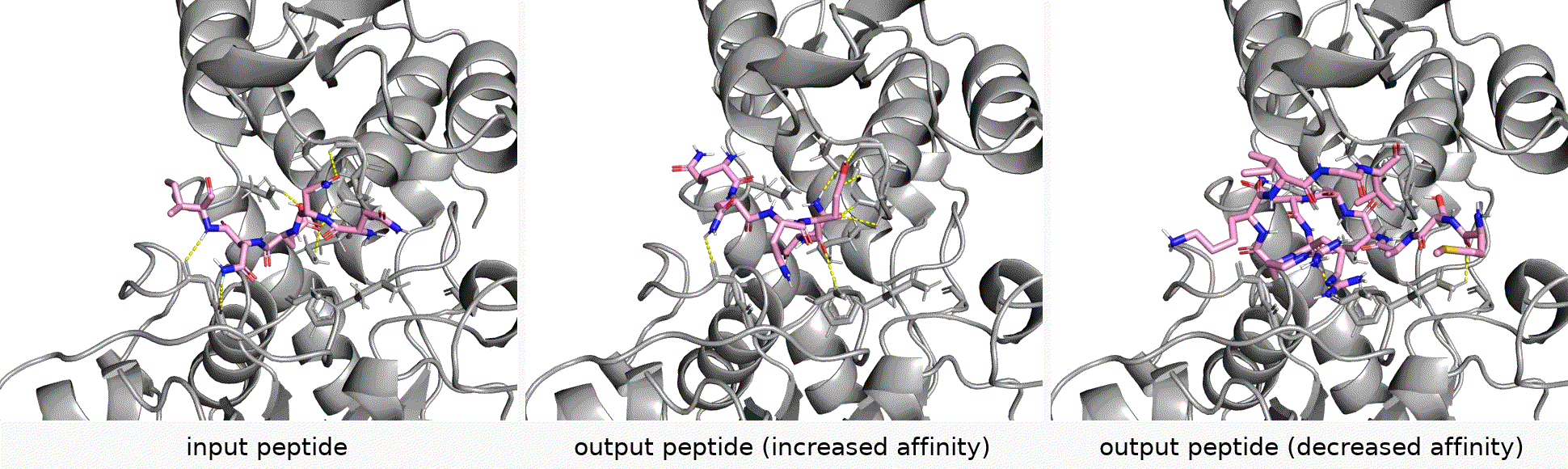Authors: Shengchao Liu, Yanjing Li, Zhuoxinran Li, Anthony Gitter, Yutao Zhu, Jiarui Lu, Zhao Xu, Weili Nie, Arvind Ramanathan, Chaowei Xiao*, Jian Tang*, Hongyu Guo*, Anima Anandkumar*
* jointly supervised
[Project Page] [ArXiv] [Datasets on HuggingFace] [Checkpoints on HuggingFace]
conda create -n ProteinDT python=3.7
conda activate ProteinDT
conda install -y numpy networkx scikit-learn
pip install torch==1.10.*
pip install transformers
pip install lxml
# for TAPE
pip install lmdb
pip install seqeval
# for baseline ChatGPT
pip install openai==0.28.0
# for baseline Galactica
pip install accelerate
# for visualization
pip install matplotlib
# for binding editing
pip install h5py
pip install torch_geometric==2.0 torch_scatter torch_sparse torch_cluster
pip install biopython
# for ESM folding
pip install "fair-esm[esmfold]"
pip install dm-tree omegaconf ml-collections einops
pip install fair-esm[esmfold]==2.0.0 --no-dependencies # Override deepspeed==0.5
pip install 'dllogger @ git+https://github.com/NVIDIA/dllogger.git'
pip install 'openfold @ git+https://github.com/aqlaboratory/openfold.git@4b41059694619831a7db195b7e0988fc4ff3a307'
conda install -c conda-forge -yq mdtraj
# for ProteinDT
pip install .
Please check folder preprocess/SwissProtCLAP for SwissProtCLAP construction from UniProt.
We also provide a copy of SwissProtCLAP at this HuggingFace link. Or you can use the following script:
from huggingface_hub import HfApi, snapshot_download
api = HfApi()
snapshot_download(repo_id="chao1224/ProteinDT", repo_type="dataset", cache_dir='./')
Then move the data under ./data folder. The data structure is
./data/
└── SwissProtCLAP
├── protein_sequence.txt
└── text_sequence.txt
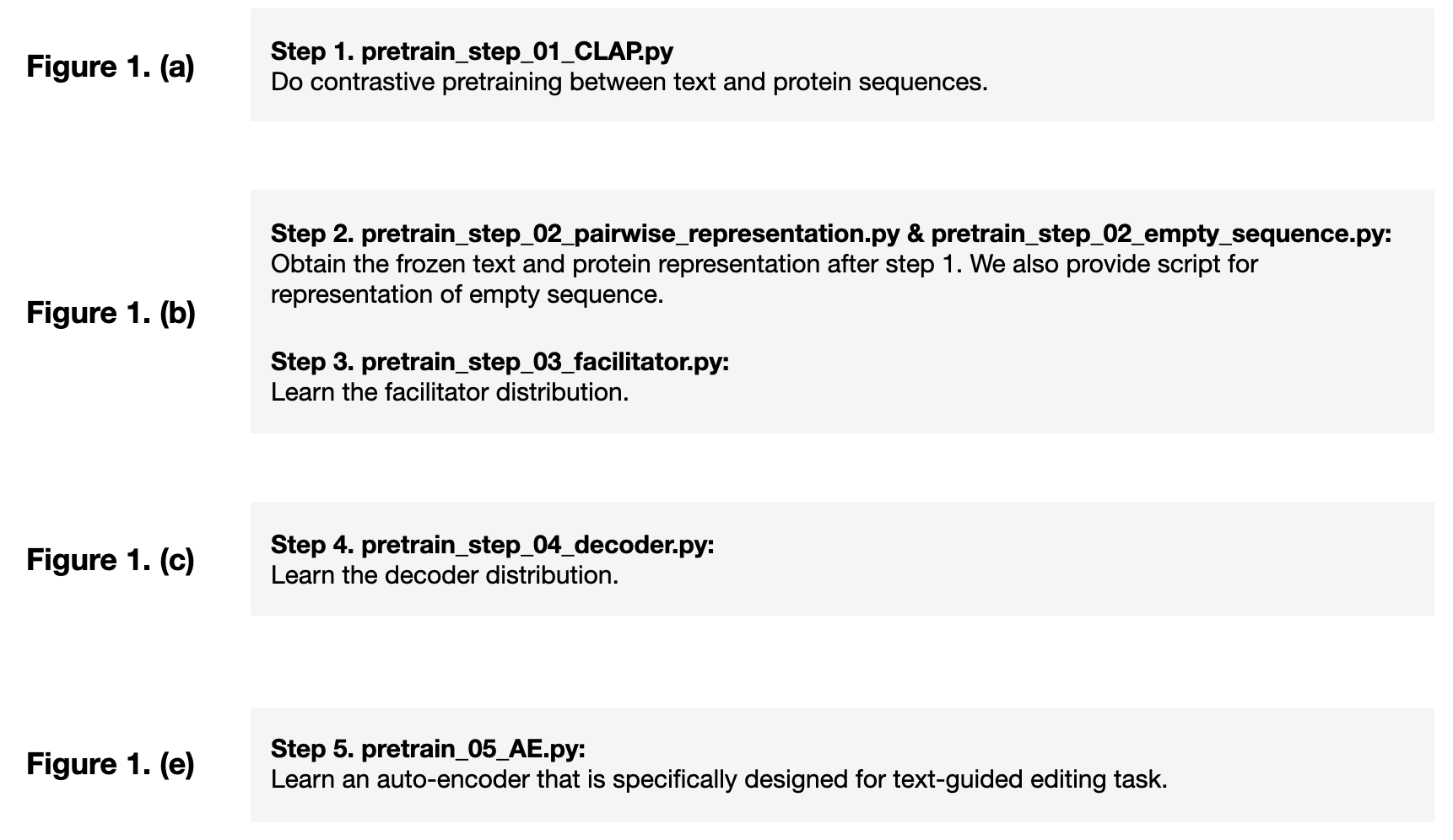
Go to folder examples, and do the pretraining in 5 steps. We summarize the logics of these 5 steps as below:
The pretrained checkpoints can be found at this HuggingFace link.
Before getting started, first we need to define our output home folder, e.g., export OUTPUT_DIR=../output/ProteinDT/hyper_01.
-
Step 1. Conduct CLAP pretraining
-
On a single GPU card:
python pretrain_step_01_CLAP.py \ --protein_lr=1e-5 --protein_lr_scale=1 \ --text_lr=1e-5 --text_lr_scale=1 \ --protein_backbone_model=ProtBERT_BFD \ --epochs=10 --batch_size=9 --num_workers=0 \ --output_model_dir="$OUTPUT_DIR" -
We also support distribution learning with DDP. Example of using a server with 8 GPU cards:
CUDA_VISIBLE_DEVICES=0,1,2,3,4,5,6,7 \ python pretrain_step_01_CLAP.py \ --protein_lr=1e-5 --protein_lr_scale=1 \ --text_lr=1e-5 --text_lr_scale=1 \ --protein_backbone_model=ProtBERT_BFD \ --epochs=10 --batch_size=9 --num_workers=0 \ --output_model_dir="$OUTPUT_DIR"
-
-
Step 2. Obtain frozen representation:
python pretrain_step_02_empty_sequence.py \ --protein_backbone_model=ProtBERT_BFD \ --batch_size=16 --num_workers=0 \ --pretrained_folder="$OUTPUT_DIR" python pretrain_step_02_pairwise_representation.py \ --protein_backbone_model=ProtBERT_BFD \ --batch_size=16 --num_workers=0 \ --pretrained_folder="$OUTPUT_DIR" -
Step 3. Learn the facilitator distribution:
python pretrain_step_03_facilitator.py \ --protein_lr=1e-5 --protein_lr_scale=1 \ --text_lr=1e-5 --text_lr_scale=1 \ --protein_backbone_model=ProtBERT_BFD \ --epochs=10 --batch_size=9 --num_workers=0 \ --pretrained_folder="$OUTPUT_DIR" \ --output_model_folder="$OUTPUT_DIR"/step_03_Gaussian_10 -
Step 4. Learn the decoder distribution. Notice that we have three types of decoder distribution models:
- A Transformer-based auto-regressive decoder. Here we adopt the T5 architecture.
python pretrain_step_04_decoder.py \ --num_workers=0 --lr=1e-4 --epochs=50 \ --decoder_distribution=T5Decoder \ --score_network_type=T5Base \ --hidden_dim=16 \ --pretrained_folder="$OUTPUT_DIR" \ --output_folder="$OUTPUT_DIR"/step_04_T5 - A discrete denoising diffusion model (multinomial diffusion).
-
Using RNN as score network:
python pretrain_step_04_decoder.py \ --num_workers=0 --lr=1e-4 --epochs=50 \ --decoder_distribution=MultinomialDiffusion \ --score_network_type=RNN \ --hidden_dim=16 \ --pretrained_folder="$OUTPUT_DIR" \ --output_folder="$OUTPUT_DIR"/step_04_MultiDiffusion_RNN -
Using BERT as score network:
python pretrain_step_04_decoder.py \ --num_workers=0 --lr=1e-4 --epochs=50 \ --decoder_distribution=MultinomialDiffusion \ --score_network_type=BertBase \ --hidden_dim=16 \ --pretrained_folder="$OUTPUT_DIR" \ --output_folder="$OUTPUT_DIR"/step_04_MultiDiffusion_BERT
-
- A Transformer-based auto-regressive decoder. Here we adopt the T5 architecture.
-
Step 5. learn an auto-encoder that is specifically designed for text-guided editing task. You can also treat this as a downstream task.
python pretrain_step_05_AE.py \ --num_workers=0 --lr=1e-4 --epochs=50 \ --pretrained_folder="$OUTPUT_DIR" \ --output_folder="$OUTPUT_DIR"/step_05
We include three types of downstream tasks, as will be introduced below. You can find the scripts for first two downstream tasks under folder scripts.
First let's go to the folder examples/downstream_Text2Protein.
Then we sample text sequences for text-to-protein generation:
python step_01_text_retrieval.py
We also provide the sampled text data in step_01_text_retrieval.txt. You can replace it with the text sequences you want to use.
Now we can do the text-to-sequence generation, e.g., if we use T5 as the decoder:
export OUTPUT_DIR=../../output/ProteinDT/hyper_01
python step_02_inference_ProteinDT.py \
--decoder_distribution=T5Decoder --score_network_type=T5Base \
--num_workers=0 --hidden_dim=16 --batch_size=8 \
--pretrained_folder="$OUTPUT_DIR" \
--step_04_folder="$OUTPUT_DIR"/step_04_T5 \
--num_repeat=16 --use_facilitator --AR_generation_mode=01 \
--output_text_file_path="$OUTPUT_DIR"/step_04_T5/downstream_Text2Protein/step_02_inference.txt
First let's go to the folder examples/downstream_Editing.
The dataset preparation can be found at examples/downstream_Edting/README.md. You can also find it on this HuggingFace link. We include three types of editing tasks: stability, structure, and peptide binding. In terms of the methods, we have two types: latent optimization and latent interpolation. The demo scripts are explained below.
- Structure / Stability:
editing_task: alpha, beta, Villin, Pin1.export OUTPUT_DIR=../../output/ProteinDT/hyper_01 python step_01_editing_latent_optimization.py \ --num_workers=0 --batch_size=8 \ --lambda_value=0.9 --num_repeat=16 --oracle_mode=text --temperature=2 \ --editing_task=alpha --text_prompt_id=101 \ --pretrained_folder="$OUTPUT_DIR" \ --step_05_folder="$OUTPUT_DIR"/step_05_AE \ --output_folder="$OUTPUT_DIR"/step_05_AE/downstream_Editing_latent_optimization/alpha_prompt_101_lambda_0.9_num_repeat_16_oracle_text_T_2 \ --output_text_file_path="$OUTPUT_DIR"/step_05_AE/downstream_Editing_latent_optimization/alpha_prompt_101_lambda_0.9_num_repeat_16_oracle_text_T_2/step_01_editing.txt python step_01_evaluate_structure.py \ --num_workers=0 --batch_size=8 --editing_task=alpha --text_prompt_id=101 \ --output_folder="$OUTPUT_DIR"/step_05_AE/downstream_Editing_latent_optimization/alpha_prompt_101_lambda_0.9_num_repeat_16_oracle_text_T_2 \ --output_text_file_path="$OUTPUT_DIR"/step_05_AE/downstream_Editing_latent_optimization/alpha_prompt_101_lambda_0.9_num_repeat_16_oracle_text_T_2/step_01_editing.txt - Peptide binding
export OUTPUT_DIR=../../output/ProteinDT/hyper_01 python step_01_editing_latent_optimization.py \ --num_workers=0 --batch_size=4 \ --lambda_value=0.9 --num_repeat=16 --oracle_mode=text --temperature=2 \ --editing_task=peptide_binding --text_prompt_id=101 \ --pretrained_folder="$OUTPUT_DIR" \ --step_05_folder="$OUTPUT_DIR"/step_05_AE \ --output_folder="$OUTPUT_DIR"/step_05_AE/downstream_Editing_latent_optimization/peptide_binding_prompt_101_lambda_0.9_num_repeat_16_oracle_text_T_2 \ --output_text_file_path="$OUTPUT_DIR"/step_05_AE/downstream_Editing_latent_optimization/peptide_binding_prompt_101_lambda_0.9_num_repeat_16_oracle_text_T_2/step_02_editing.txt
Notice that for latent interpolation, we have three models: auto-regressive (T5), denoising diffusion model (RNN and BERT). We provide demos scripts using T5.
- Structure / Stability:
editing_task: alpha, beta, Villin, Pin1.export OUTPUT_DIR=../../output/ProteinDT/hyper_01 python step_01_editing_latent_interpolation.py \ --editing_task=alpha --text_prompt_id=101 \ --decoder_distribution=T5Decoder --score_network_type=T5Base \ --num_workers=0 --hidden_dim=16 --batch_size=2 \ --theta=0.9 --num_repeat=16 --oracle_mode=text --AR_generation_mode=01 --AR_condition_mode=expanded \ --pretrained_folder="$OUTPUT_DIR" --step_04_folder="$OUTPUT_DIR"/step_04_T5 \ --output_folder="$OUTPUT_DIR"/step_04_T5/downstream_Editing_latent_interpolation_alpha/prompt_101_theta_0.9_num_repeat_16_oracle_text_inference_01_expanded \ --output_text_file_path="$OUTPUT_DIR"/step_04_T5/downstream_Editing_latent_interpolation_alpha/prompt_101_theta_0.9_num_repeat_16_oracle_text_inference_01_expanded/step_01_editing.txt python step_01_evaluate_structure.py \ --num_workers=0 --batch_size=1 \ --editing_task=alpha --text_prompt_id=101 \ --output_folder="$OUTPUT_DIR"/step_04_T5/downstream_Editing_latent_interpolation_alpha/prompt_101_theta_0.9_num_repeat_16_oracle_text_inference_01_expanded \ --output_text_file_path="$OUTPUT_DIR"/step_04_T5/downstream_Editing_latent_interpolation_alpha/prompt_101_theta_0.9_num_repeat_16_oracle_text_inference_01_expanded/step_01_editing.txt - Peptide binding
export OUTPUT_DIR=../../output/ProteinDT/hyper_01 python step_02_binding_editing_latent_interpolation.py \ --editing_task=peptide_binding --text_prompt_id=101 \ --decoder_distribution=T5Decoder --score_network_type=T5Base \ --num_workers=0 --hidden_dim=16 --batch_size=1 \ --theta=0.9 --num_repeat=16 --oracle_mode=text --AR_generation_mode=01 --AR_condition_mode=expanded \ --pretrained_folder="$OUTPUT_DIR" --step_04_folder="$OUTPUT_DIR"/step_04_T5 \ --output_folder="$OUTPUT_DIR"/step_04_T5/downstream_Editing_latent_interpolation_peptide_binding/prompt_101_theta_0.9_num_repeat_16_oracle_text_inference_01_expanded \ --output_text_file_path="$OUTPUT_DIR"/step_04_T5/downstream_Editing_latent_interpolation_peptide_binding/prompt_101_theta_0.9_num_repeat_16_oracle_text_inference_01_expanded/step_02_editing.txt
First please download the TAPE data following instructions here. We also provide it at this HuggingFace link.
Under examples, and the script is downstream_TAPE.py. We follow the exactly same hyper-parameter as OntoProtein.
python downstream_TAPE.py \
--task_name=ss3 \
--seed=3 \
--learning_rate=3e-5 \
--num_train_epochs=5 \
--per_device_train_batch_size=2 \
--gradient_accumulation_steps=8 \
--warmup_ratio=0.08 \
--pretrained_model=ProteinDT \
--pretrained_folder="$OUTPUT_DIR" \
--output_dir="$OUTPUT_DIR"/downstream_TAPE
Feel free to cite this work if you find it useful to you!
@article{liu2023text,
title={A Text-guided Protein Design Framework},
author={Shengchao Liu, Yanjing Li, Zhuoxinran Li, Anthony Gitter, Yutao Zhu, Jiarui Lu, Zhao Xu, Weili Nie, Arvind Ramanathan, Chaowei Xiao, Jian Tang, Hongyu Guo, Anima Anandkumar},
journal={arXiv preprint arXiv:2302.04611},
year={2023}
}