Meng Lu1, Charles N. Christensen1,*, Jana M. Weber1,*, Tasuku Konno, Nino F. Läubli1, Katharina M. Scherer1, Edward Avezov, Pietro Lio, Alexei A. Lapkin, Gabriele S. Kaminski Schierle1, Clemens F. Kaminski1
1University of Cambridge, Department of Chemical Engineering and Biotechnology
*Author of this repository - GitHub username: charlesnchr - Email address: charles.n.chr@gmail.com
Pre-print: https://www.biorxiv.org/content/10.1101/2022.05.17.492189v1
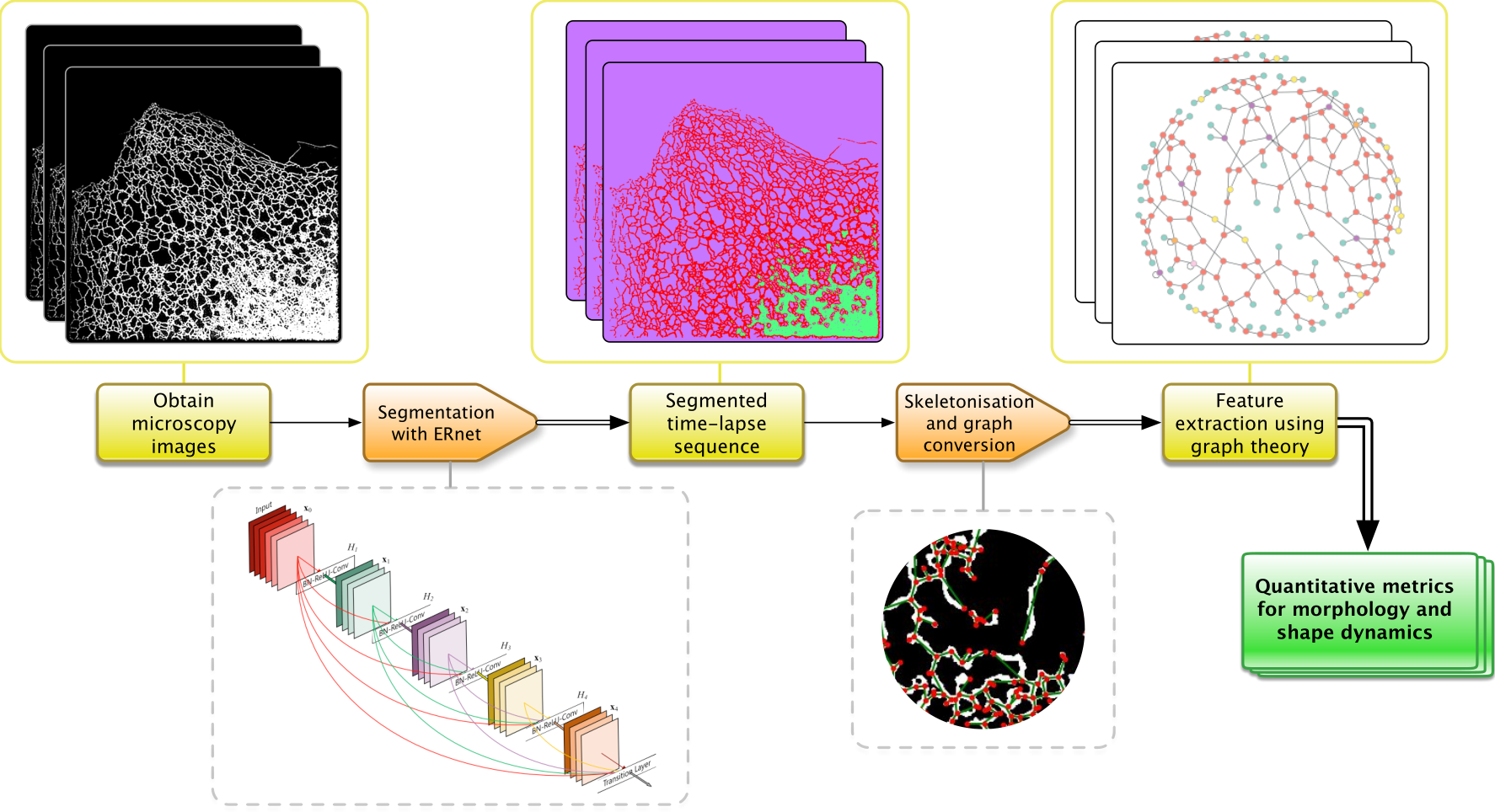
ERnet is a vision transformer-based processing pipeline for segmentation and analysis of temporal image sequences of the endoplasmic reticulum. The main component is the vision transformer model used for segmentation, inspired by Swin. Other steps in the pipeline that facilitates quantitative analysis are shown in the figure above.
The method follows our previous work, in which we used a residual neural network for two-class image segmentation, ERnet — Residual network.
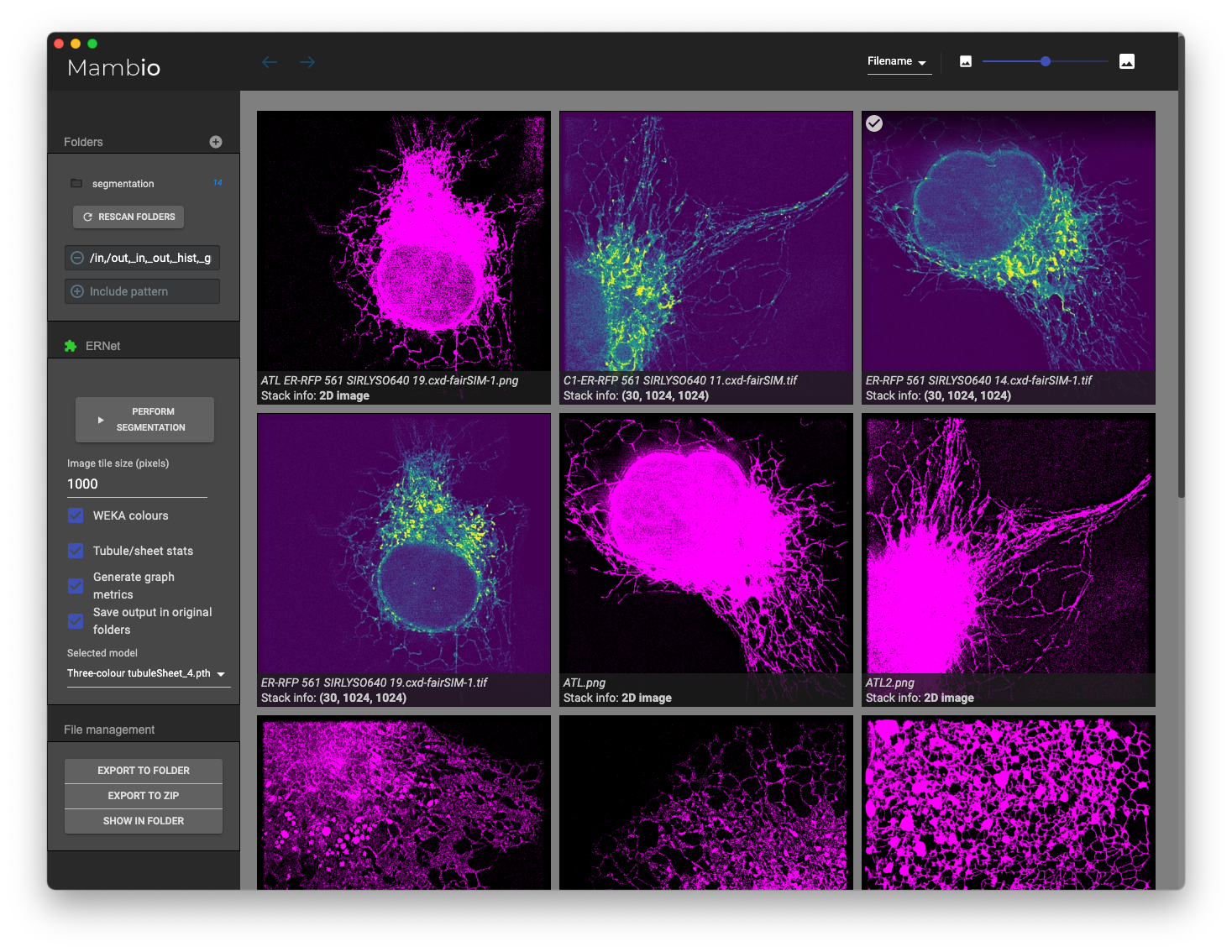
Get started right away with our Colab notebooks. If you prefer running the code locally, we provide Jupyter notebooks and Python scripts in this repository. Another option for running ERnet is to use it as a plugin in the graphical front-end, Mambio, based on an integration of the Electron UI framework and a Python environment. Mambio is also included in this repository.
The graphical user interface, Mambio, is provided in the folder Graphical-App. Instructions and further information can be found in the corresponding README.
Code for training models based on the ERnet architecture can be found in the folder Training, and further information is also in the README file of that folder. A script that generates synthetic ER images can be found in Training/Simulate_ER, see the Synthetic data generation pipeline section.
To run inference with ERnet locally, we provide the graphical user interface Mambio: Multi-purpose Advanced ML-based Batch Image Operations. Mambio is a more general software that extends beyond image segmentation — there is also a plugin for ML-SIM.
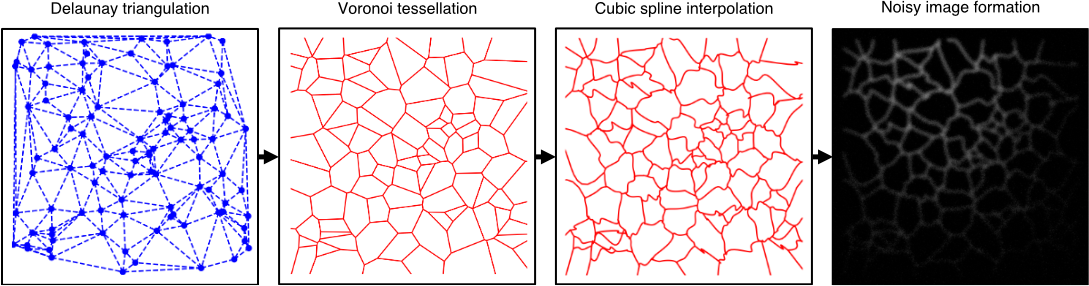
We propose a model for synthesising ground truth ER images with a simple algorithm. First a set of uniformly distributed coordinates are generated. Delaunay triangulation is performed on the coordinates providing a set of triangles. The circumcentre of each triangle becomes a vertex in a Voronoi diagram. The diagram is formed by connecting vertices between adjacent triangles. This procedure is called Voronoi tessellation. Each edge in the undirected graph corresponding to the Voronoi diagram is the perturbed in the following way: the midpoint of an edge is moved in a random direction by a random distance that is smaller than the length of the edge. The displaced midpoint and the two ends of the edge are then used the produce a smooth curve using cubic spline interpolation of the three points. Finally, the network of vertices connected with interpolated curves are processed with a image formation model that applies blur (diffraction) and noise (Poisson noise and Gaussian read-out noise).
The steps of this algorithm are illustrated on the figure above. The implementation can be found in Training/Simulate_ER, and the script can be used to generate a collection of test images with
python Simulate_ER_images_script.py
Parameters are given in the file.
ERNet — Residual network
Segmentation of Endoplasmic Reticulum microscopy images using modified CNN-based image restoration models.
Meng Lu1, Francesca W. van Tartwijk1, Julie Qiaojin Lin1, Wilco Nijenhuis, Pierre Parutto, Marcus Fantham1, Charles N. Christensen1,*, Edward Avezov, Christine E. Holt, Alan Tunnacliffe, David Holcman, Lukas C. Kapitein, Gabriele Kaminski Schierle1, Clemens F. Kaminski1
1University of Cambridge, Department of Chemical Engineering and Biotechnology
*Author of this repository - GitHub username: charlesnchr
Pre-print: https://www.biorxiv.org/content/10.1101/2020.01.15.907444v2
Paper: https://www.science.org/doi/10.1126/sciadv.abc7209