ViewBS has several top level commands which determine the required and optimal arguments. These top level commands can be divided into two parts: methylation report and data visualization of functional regions.
Methylation report part has several different top commands which can generate report about read coverage, distribution of methylation level, global methylation leve, etc.
The part of visualization for functional regions also has several different top commands. For ViewBS, the first input that users should provide is the regions of interest. These regions could be functional elements, like genes, transposable elements (TE), or differentially methylated regions (DMR). The other type of input that the users should provide is the methylation information. Methylation information are the outputs from BS-seq aligner, like Bismark, etc.
Here is the workflow of ViewBS:
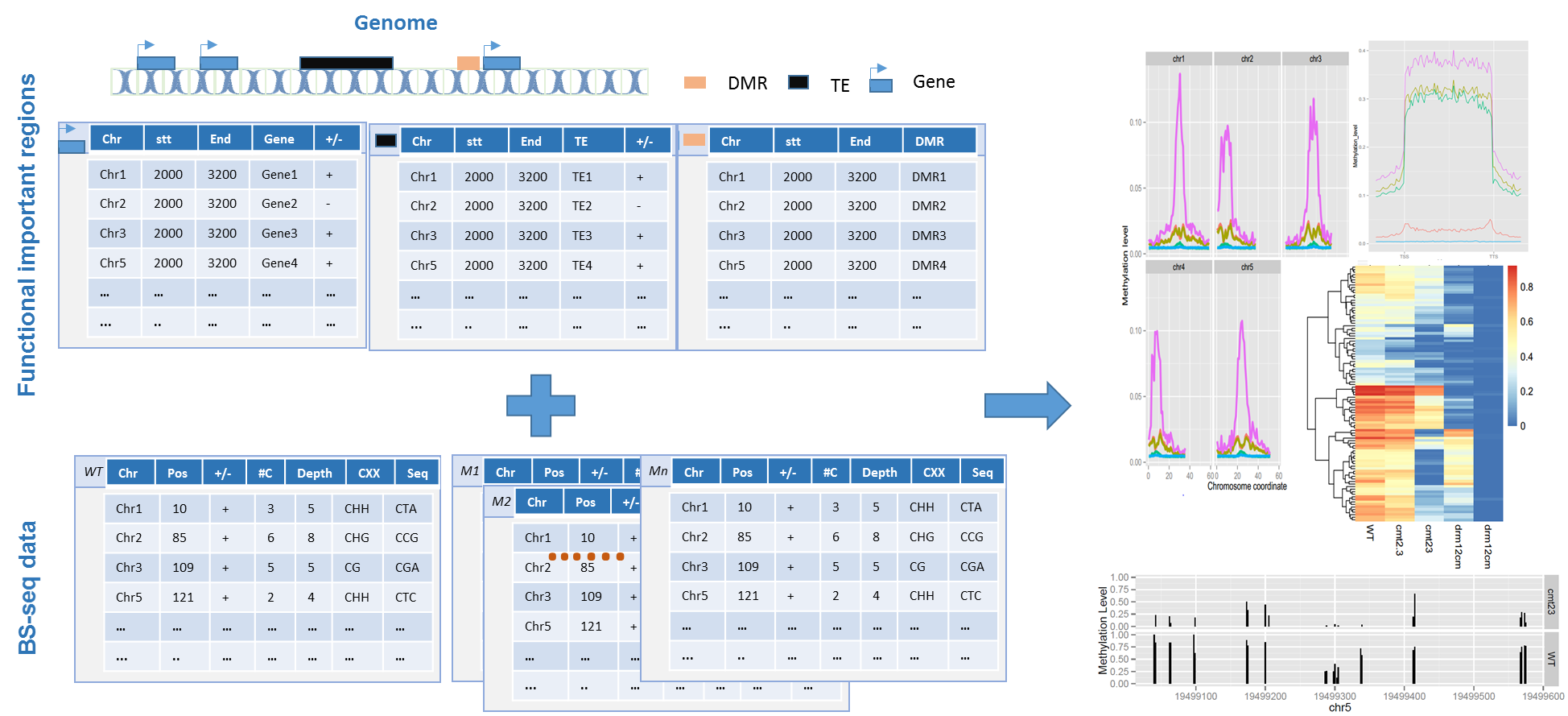 The workflow of ViewBS commands
The workflow of ViewBS commands
To make the installation of dependencies easier, a script was developped.
perl INSTALL.plcan be used as a helper to install and check the dependencies.
-
Install htslib
-
Perl version: >= 5.14.4
-
Perl packages:
- Getopt::Long::Subcommand - Process command-line options, with subcommands and completion
- Bio::DB::HTS::Tabix - Object oriented access to the underlying tbx C methods
- Bio::SeqIO - Handler for SeqIO Formats
wget https://raw.githubusercontent.com/xie186/ViewBS/master/ext_tools/cpanm chmod 755 cpanm cpanm --local-lib=~/perl5 local::lib && eval $(perl -I ~/perl5/lib/perl5/ -Mlocal::lib) ./cpanm Getopt::Long::Subcommand ./cpanm Bio::DB::HTS::Tabix ./cpanm Bio::SeqIO -
R version: > 3.3.0
-
R packages
- ggplot2
- pheatmap
- reshape2
- cowplot
Install the required libraries in R:
install.packages("ggplot2", dep=T) install.packages("cowplot", dep=T) install.packages("pheatmap", dep=T) install.packages("reshape2", dep=T)
- Input file: Genome-wide cytosine methylation report
ViewBS uses Genome-wide cytosine methylation report as input file. It is sorted by chromosomal coordinates but also contains the sequence context and is in the following format:
<chromosome> <position> <strand> <count methylated> <count unmethylated> <C-context> <trinucleotide context>
Please see details in Bismark websites.
Tips: how to generate Genome-wide Cytosine Methylation Report
If you already have finished the mapping using Bismark, you should have a sam/bam file. Let's say you have a sam file named test.sam. What you can do to generate Genome-wide Cytosine Methylation Report is:
### This step will generate several files: bismark_methylation_extractor --bedGraph --CX test.sam ### This step will generate a file named bis_test.tab coverage2cytosine -CX -o test.bis_rep.cov --genome_folder ara/ test.bismark.cov
*For BS-seq that is processed by Bismark but by other tools like BRAT, BS seeker2, ViewBS provides supports to convert DNA methylation data in other format to the format of genome-wide cytosine methylation report. Supports for other tools will be developed upon requests from the users. If you have DNA methylation data generated by other tools and you have difficulties on converting the data format, just give a post in the issuse. We're happy to add new functions for the file format conversion. *
For details, please see the link below: https://github.com/xie186/ViewBS/wiki/Support-for-nonBismark-results
- Tabix indexing
Since ViewBS uses Bio::DB::HTS::Tabix to quickly retrieves information from the input (TAB-delited) files, the Genome-wide Cytosine Methylation Report files should be bgzipped and tabix indexed. bgzip and tabix .
Note: tabix and bgzip binaries are now part of the HTSlib project. https://github.com/samtools/htslib
Here is an example:
bgzip test.bis_rep.cov ## test.bis_rep.cov.gz will be generated. Note: test.bis_rep.cov shoud be sorted based on chromosome coordinates.
tabix -p vcf test.bis_rep.cov.gz ## test.bis_rep.cov.gz.tbi will be generated. Now test.bis_rep.cov.gz can be used as input for ViewBS.
https://gitlab.com/BS-seq/ViewBS_testdata
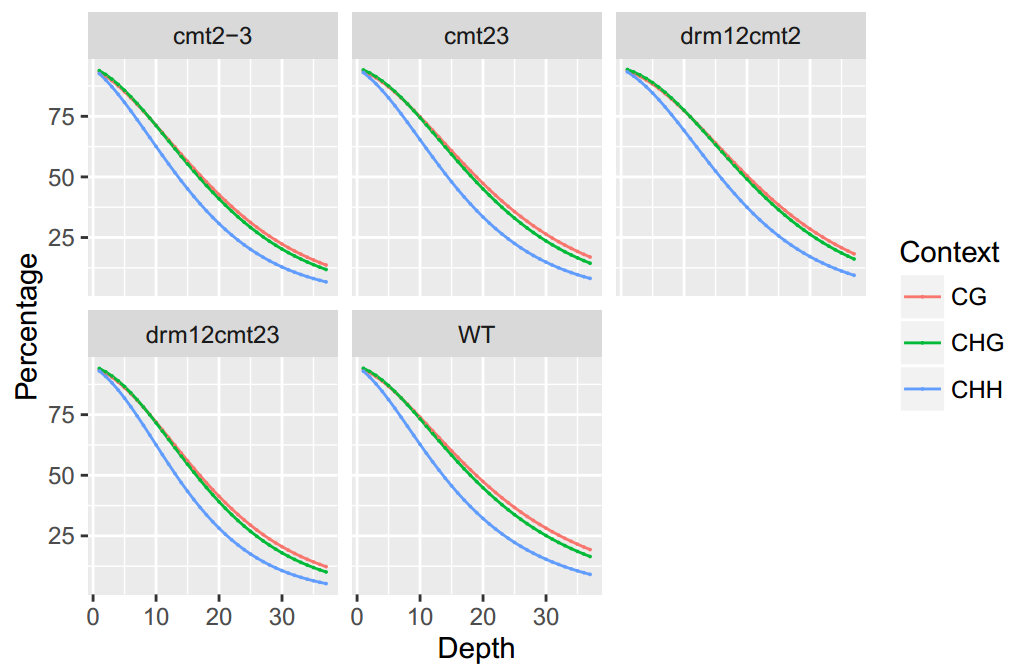
An Example of Coverage Distribution of BS-seq
To generate the figure above, use the command shown as below:
ViewBS MethCoverage --reference TAIR10_chr_all.fasta --sample bis_WT.tab.gz,WT --sample bis_cmt23.tab.gz,cmt23 --sample bis_cmt2-3.tab.gz,cmt2-3 --sample bis_drm12cmt23.tab.gz,drm12cmt12 --sample bis_drm12cmt2.tab.gz,drm12cmt2 --outdir methCoverage --prefix cmt2_proj_allsam
Under methCoverage folder, there will be three files generated.
- Table for global methylation level.
| Sample | Context | Depth | Percentage |
|---|---|---|---|
| cmt2-3 | CG | 1 | 93.3323115145888 |
| cmt2-3 | CG | 2 | 91.6474703919394 |
| ... | ... | ... | ... |
| ... | ... | ... | ... |
| WT | CG | 1 | 93.8364493009668 |
- A shell script which can re-generate the figure in PDF file.
- A figure in PDF file.
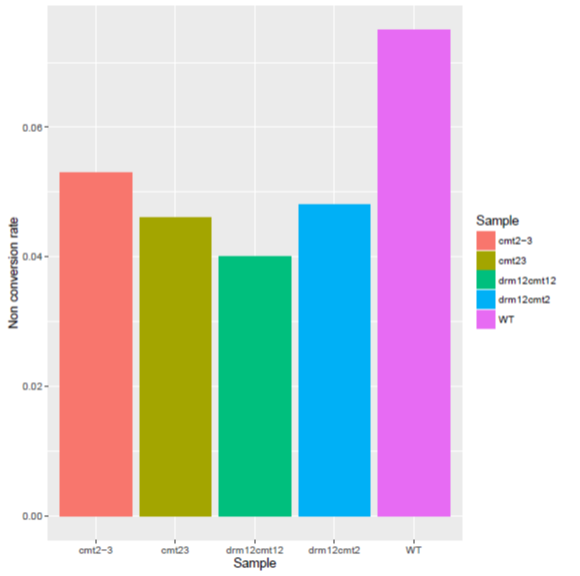
An Example of BisNonConvRate
To generate the figure above, use the command shown as below:
ViewBS BisNonConvRate --chrom chrC --sample bis_WT.tab.gz,WT --sample bis_cmt23.tab.gz,cmt23 --sample bis_cmt2-3.tab.gz,cmt2-3 --sample bis_drm12cmt2.tab.gz,drm12cmt2 --sample bis_drm12cmt23.tab.gz,drm12cmt23 --outdir BisNonConvRate --prefix cmt2_proj_allsam
Under BisNonConvRate, there will be three files generated.
- Table for global methylation level.
| Sample | BisNonConvRate |
|---|---|
| cmt2-3 | 0.053 |
| drm12cmt2 | 0.048 |
| drm12cmt12 | 0.040 |
| cmt23 | 0.046 |
| WT | 0.075 |
- A shell script which can re-generate the figure in PDF file.
- A figure in PDF file.
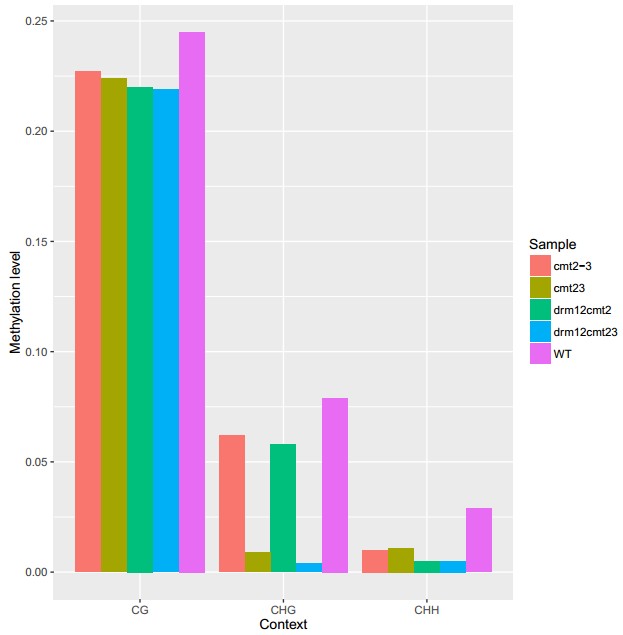
An Example of GlobalMethLev
To generate the figure above, use the command shown as below:
ViewBS GlobalMethLev --sample bis_WT.tab.gz,WT --sample bis_cmt23.tab.gz,cmt23 --sample bis_cmt2-3.tab.gz,cmt2-3 --sample bis_drm12cmt2.tab.gz,drm12cmt2 --sample bis_drm12cmt23.tab.gz,drm12cmt23 --outdir methGlobal --prefix cmt2_proj_allsam
Under methGlobal, there will be three files generated.
- Table for global methylation level.
| Sample | CG | CHG | CHH |
|---|---|---|---|
| cmt2-3 | 0.227 | 0.062 | 0.010 |
| drm12cmt2 | 0.220 | 0.058 | 0.005 |
| cmt23 | 0.224 | 0.009 | 0.011 |
| drm12cmt23 | 0.219 | 0.004 | 0.005 |
| WT | 0.245 | 0.079 | 0.029 |
- A shell script which can re-generate the figure in PDF file.
- A figure in PDF file.
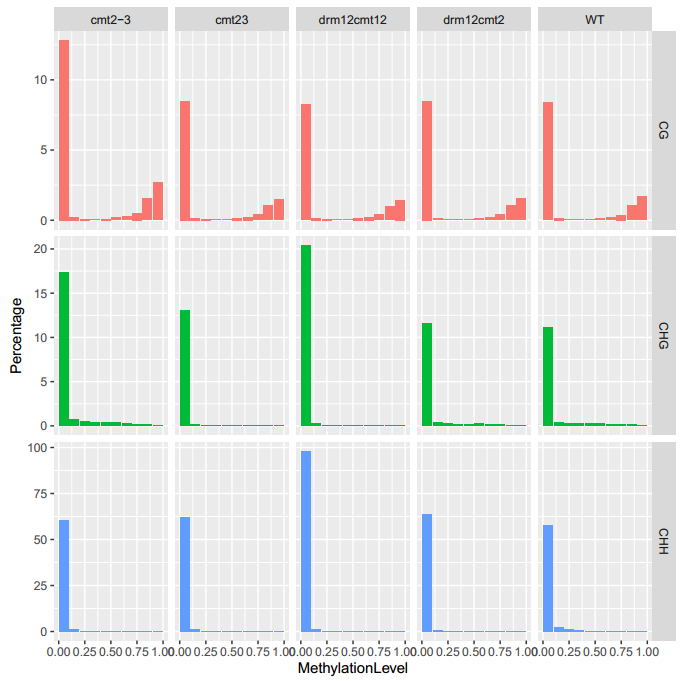
An Example of MethLevDist
To generate the figure above, use the command shown as below:
ViewBS.pl MethLevDist --sample bis_WT.tab.gz,WT --sample bis_cmt23.tab.gz,cmt23 --sample bis_cmt2-3.tab.gz,cmt2-3 --sample bis_drm12cmt23.tab.gz,drm12cmt12 --sample bis_drm12cmt2.tab.gz,drm12cmt2 --outdir methLevDist --prefix cmt2_proj_allsam --binMethLev 0.1
- Table for numbers and percentages of sites in each methylation level bin.
| Sample | Context | MethLevBinMidPoint | Number | Percentage |
|---|---|---|---|---|
| cmt2-3 | CG | 0.05 | 3305969 | 12.83 |
| cmt2-3 | CG | 0.15 | 62823 | 0.24 |
| cmt2-3 | CG | 0.25 | 25182 | 0.09 |
| ... | ... | ... | ... | .. |
| WT | CG | 0.05 | 3470693 | 13.73 |
- A shell script which can re-generate the figure in PDF file.
- A figure in PDF file.
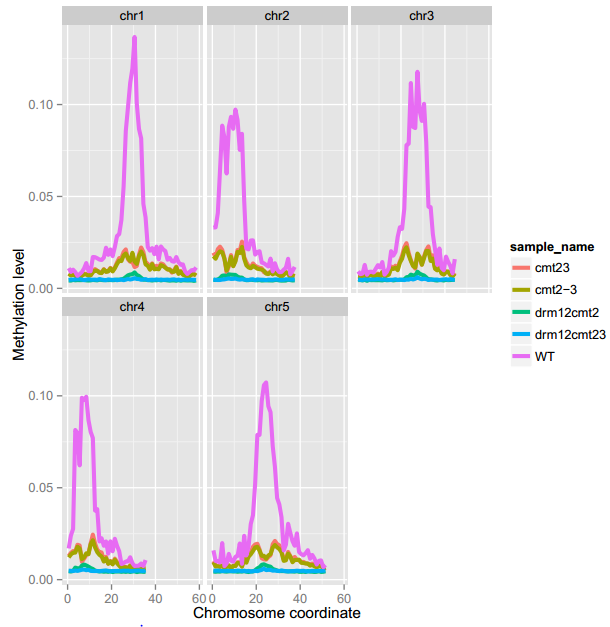
An example of MethGeno
To generate the figure above, use the command shown as below:
ViewBS MethGeno --genomeLength TAIR10_chr_all.fasta.fai --sample bis_WT.tab.gz,WT --sample bis_cmt23.tab.gz,cmt23 --sample bis_cmt2-3.tab.gz,cmt2-3 --sample bis_drm12cmt2.tab.gz,drm12cmt2 --sample bis_drm12cmt23.tab.gz,drm12cmt23 --prefix bis_geno_sample --context CHH
Note: fai file can generated by samtools: samtools faidx TAIR10_chr_all.fasta
Region file format:
- 1st column: chromsome ID;
- 2nd column: start position;
- 3rd column: end position;
- 4th column: region ID
Note: If the file has 4th column, each row in this column should be unique.
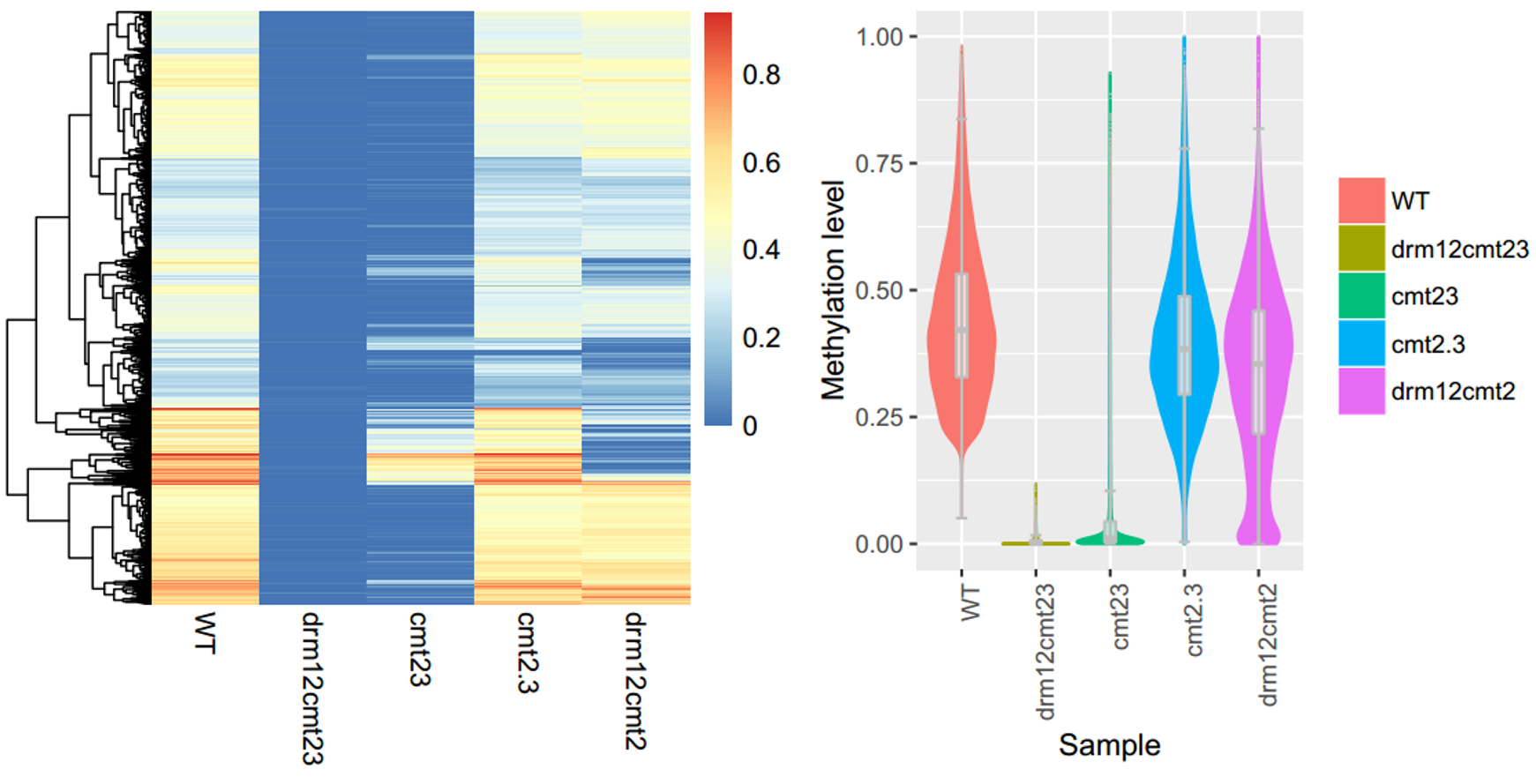
An example of MethHeatmap
To generate the figure above, use the command shown as below:
ViewBS MethOverRegion --region TAIR10_Transposable_Elements.chr1.bed --sample bis_WT.tab.gz,WT --sample bis_cmt23.tab.gz,cmt23 --sample bis_cmt2-3.tab.gz,cmt2-3 --sample bis_drm12cmt2.tab.gz,drm12cmt2 --sample bis_drm12cmt23.tab.gz,drm12cmt23 --prefix bis_TE_chr1_sample --context CHG
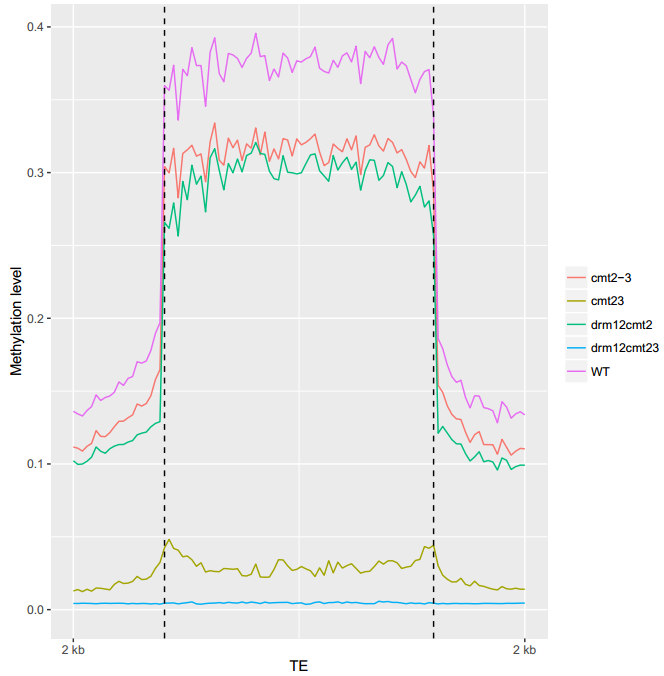
An example of MethOverregion
ViewBS MethOverRegion --region TAIR10_Transposable_Elements.chr1.bed --sample bis_WT.tab.gz,WT --sample bis_cmt23.tab.gz,cmt23 --sample bis_cmt2-3.tab.gz,cmt2-3 --sample bis_drm12cmt2.tab.gz,drm12cmt2 --sample bis_drm12cmt23.tab.gz,drm12cmt23 --prefix bis_TE_chr1_sample --context CHG
Besides providing sample and region information in the commind line, you can also read the information from a TEXT file. For example, if you are interested in more than one group of genes and you want to study the differences of DNA methylation patterns in the one sample, the methylation information can also be read from a TEXT file. Instead of giving an explicit sample information pairs, you need to write "file:" followed by the name of the TEXT file. In this case, you can only use --sample once and you cann't use --region anymore.
The TEXT file should follow the following format:
| #MethReportFile | LegendName | RegionFile |
|---|---|---|
| DNAmethylation | RegionName1 | Region_file2 |
Here is an example:
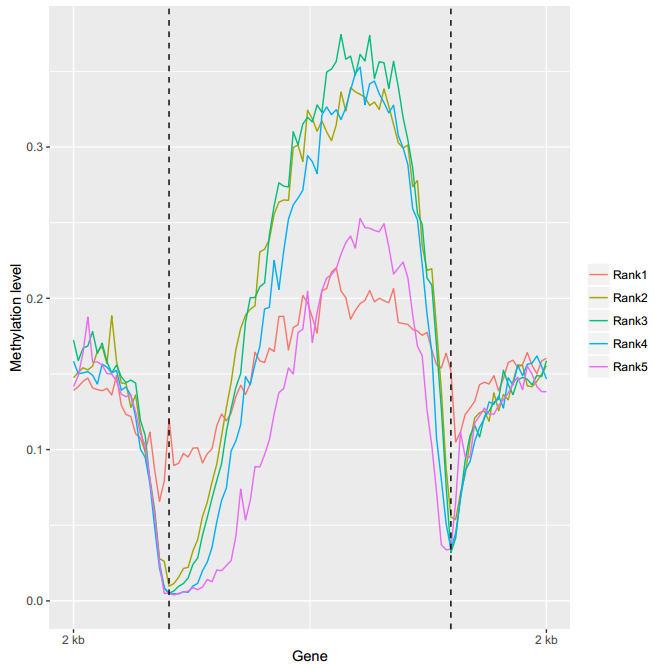
ViewBS MethOverRegion --sample file:sampl_info_tab.txt --prefix bis_gene_5rank --context CG --outdir MethOverRegion
The genes were devided into quintiles based on gene expression level. Rank1 group was the group with lowest expression level. Users can use this method to study the correlation between DNA methylation and gene expression.
| #DNAmethylationData | Region | RegionFile |
|---|---|---|
| bis_WT.tab.gz | Rank1 | TAIR10_GFF3_genes.WT.rank1.tab |
| bis_WT.tab.gz | Rank2 | TAIR10_GFF3_genes.WT.rank2.tab |
| bis_WT.tab.gz | Rank3 | TAIR10_GFF3_genes.WT.rank3.tab |
| bis_WT.tab.gz | Rank4 | TAIR10_GFF3_genes.WT.rank4.tab |
| bis_WT.tab.gz | Rank5 | TAIR10_GFF3_genes.WT.rank5.tab |
Here is the figure generated by the command line above:
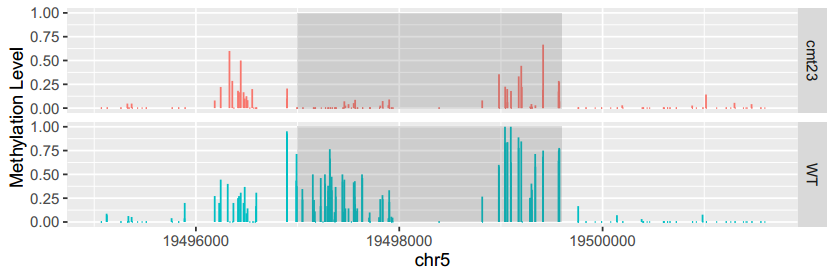
View MethOneRegion will output the methylation information for one region give by the users and then plot the methylation levels across the chromsomesome region.
Here is an example:
To generate the figure above, you can use the following command line:
ViewBS MethOneRegion --region chr5:19499001-19499600 --sample bis_WT.tab.gz,WT --sample bis_cmt23.tab.gz,cmt23 --prefix chr5_19499001-19499600 --context CHG
If you have bugs, feature requests, please report the issues here: (https://github.com/readbio/ViewBS/issues).
ViewBS uses GNU GPLv3 and is free for use by academic users. If you want to use it in commercial settings, please contact us.
Currently the manuscript of ViewBS is in preparation.
- Nanjing Agricultural University
Drs. Xiaosan Huang (huangxs@njau.edu.cn), Kong-Qing Li (likq@njau.edu.cn) and Shaoling Zhang (slzhang@njau.edu.cn).
- Purdue Univeristy
Drs. Shaojun Xie: (Email: xie186@purdue.edu) and Jyothi Thimmapuram (jyothit@purdue.edu)