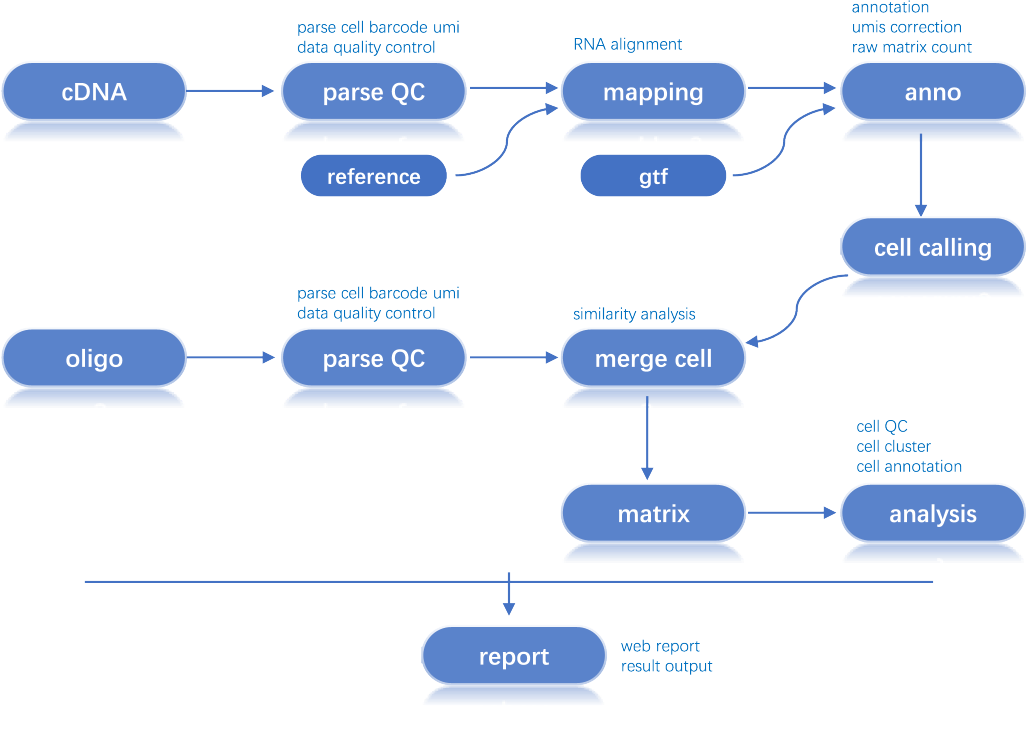
An open source and flexible pipeline to analyze high-throughput DNBelab C SeriesTM single-cell RNA datasets.
Hardware/Software requirements
- x86-64 compatible processors.
- require at least 50GB of RAM and 4 CPU.
- centos 7.x 64-bit operating system (Linux kernel 3.10.0, compatible with higher software and hardware configuration).
Workflow