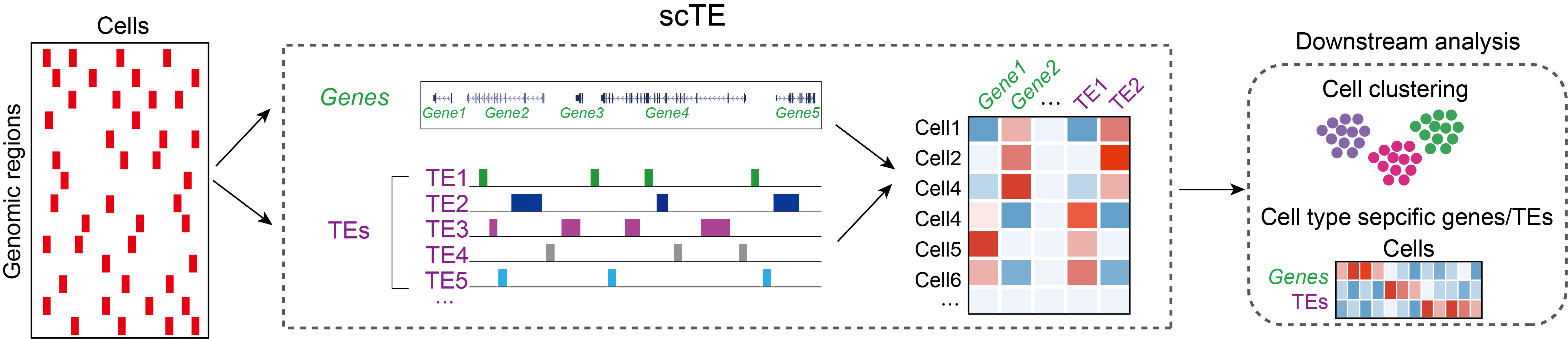
scTE takes as input:
- Aligned sequence reads (BAM/SAM format)
- The genomic location of TEs (BED format)
- The genomic location of genes (GTF format)
scTE works with python >=3.6.
$ git clone https://github.com/jphe/scTE.git
$ cd scTE
$ python setup.py installBuilding genome indices
scTE builds genome indices for the fast alignment of reads to genes and TEs. These indices can be automatically generated using the commands:
$ scTE_build -g mm10 # mouse genome
$ scTE_build -g hg38 # human genomeThese two scripts will automatically download the genome annotations, for mouse:
$ ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_mouse/release_M21/gencode.vM21.annotation.gtf.gz
$ http://hgdownload.soe.ucsc.edu/goldenPath/mm10/database/rmsk.txt.gzOr for human:
$ ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_30/gencode.v30.annotation.gtf.gz
$ http://hgdownload.soe.ucsc.edu/goldenPath/hg38/database/rmsk.txt.gzmm10, hg38 is the genome assembly version, scTE currently support mm10 and hg38.
If you want to use your customs reference, you can use the -gene -te options:
scTE_build -te TEs.bed -gene Genes.gtf -o custome.idx
-te
Six columns bed file for transposable elements annotation.
-gene
Gtf file for genes annotation.
For more informat about BED and GTF format, see from UCSC.
These annotations are then processed and converted into genome indices. The scTE algorithm will allocate
reads first to gene exons, and then to TEs by default. Hence TEs inside exon/UTR regions of genes annotated
in GENCODE will only contribute to the gene, and not to the TE score. This feature can be changed by
setting –mode/-m exclusive in scTE, which will instruct scTE to assign the reads to both TEs and genes
if a read comes from a TE inside exon/UTR regions of genes.
Analysis of 10x style scRNA-seq data
scTE makes BAM/SAM file as input, highly recommend to use unfiltered alignment file as input.
For bam file, the cell barcodes and UMI need to be integrated into the read 'CR:Z' or 'UR:Z' tage as bellow:
$ samtools view test.bam
A00269:12:H7YF2DMXX:2 0 chr10 55902580 255 50M * 0 0 GTTCTCTCCGTATGTGAGCATGGGAGATACATCCCAGAAAGGCAGAAGGG FFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFF NH:i:1 HI:i:1 AS:i:49 nM:i:0 CR:Z:CTAGAGTGTTTCGCTC CY:Z:FFFFFFFFFFFFFFFF UR:Z:TACATGACGC UY:Z:FFFFFFFFFF
A00269:13:H7YF2DMXX:2 0 chr10 55902784 255 50M * 0 0 ATAATCTTTGAGATCTCTGGTGAAAATAAGTAGCATAAAGGACAGAATCA FFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFF NH:i:1 HI:i:1 AS:i:49 nM:i:0 CR:Z:CTAGAGTGTTTCGCTC CY:Z:FFFFFFFFFFFFFFFF UR:Z:TACATGACGC UY:Z:FFFFFFFFFF
A00269:14:H7YF2DMXX:2 0 chr13 67837311 255 50M * 0 0 CTGTTCATTATTTGAGGAAATCAGGACAGGAAATCAAACATGGCAGAATC FFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFF NH:i:1 HI:i:1 AS:i:49 nM:i:0 CR:Z:ATCGAGTGTTTCGCTC CY:Z:FFFFFFFFFFFFFFFF UR:Z:TACATGACGC UY:Z:FFFFFFFFFF
A00269:15:H7YF2DMXX:2 0 chr14 114380523 255 50M * 0 0 GATCCAGATTAATTGAGACTGTTGATCCTCCTACAGGGTCGCCCTTCTCC FFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFF NH:i:1 HI:i:1 AS:i:49 nM:i:0 CR:Z:CTAGAGTGTTTCGCTC CY:Z:FFFFFFFFFFFFFFFF UR:Z:TACATGACGC UY:Z:FFFFFFFFFF
$ scTE -i inp.bam -o out -g mm10 -x mm10.exclusive.idx --hdf5 True
-i
Input file: BAM/SAM file from CellRanger or STARsolo
-o
Output file prefix
-g
"hg38" for human, "mm10" for mouse
-x
The filename of the index for the reference genome annotation generated by scTE_build
--hdf5
Save the output as .h5ad formatted file instead of csv file. Default: FalsescTE is most tuned to STARsolo or the Cell Ranger pipeline outputs, and can accept BAM files produced by either of these two programs. For other aligners, the barcode should be stored in the ‘CR:Z’ tag, and the UMI in the ‘UR:Z’ tag in the BAM file
Analysis of C1 style scRNA-seq data
If the UMI is missing or not used in the scRNA-seq technology (for example on the Fluidigm C1 platform), it can be disabled with –UMI False
(the default is True) switch in scTE. If the barcode is missing it can be disabled with the –CB False (the default is True),
and instead the cell barcodes will be taken from the names of the BAM files.
$ scTE -i inp.bam -o out -g mm10 -x mm10.exclusive.idx -CB False -UMI Falsemultiple BAM files can be provided to scTE with the –i option
$ scTE -i *.bam -o out -g mm10 -x mm10.exclusive.idx -CB False -UMI False
or
$ scTE -i input1.bam,input2.bam,... -o out -g mm10 -x mm10.exclusive.idx -CB False -UMI False
scTE is most tuned to STARsolo or the Cell Ranger pipeline outputs, and can accept BAM files produced by either of these two programs. For other aligners, the barcode should be stored in the ‘CR:Z’ tag, and the UMI in the ‘UR:Z’ tag in the BAM file
Analysis of scATAC-seq data
The genome indices were prebuilt using:
$ wget -c http://hgdownload.soe.ucsc.edu/goldenPath/mm10/database/rmsk.txt.gz -O mm10.te.txt.gz
$ zcat mm10.te.txt.gz | grep -E 'LINE|SINE|LTR|Retroposon' | cut -f6-8,11 >mm10.te.bed
$ scTEATAC_build -g mm10.te.bed -o mm10.te.atac
Then the bam file can processe using scTE with the command:
scTE_scatacseq -i input.bam -x mm10.te.atac.idx
Citation
If scTE is useful for your research, consider citing our paper.