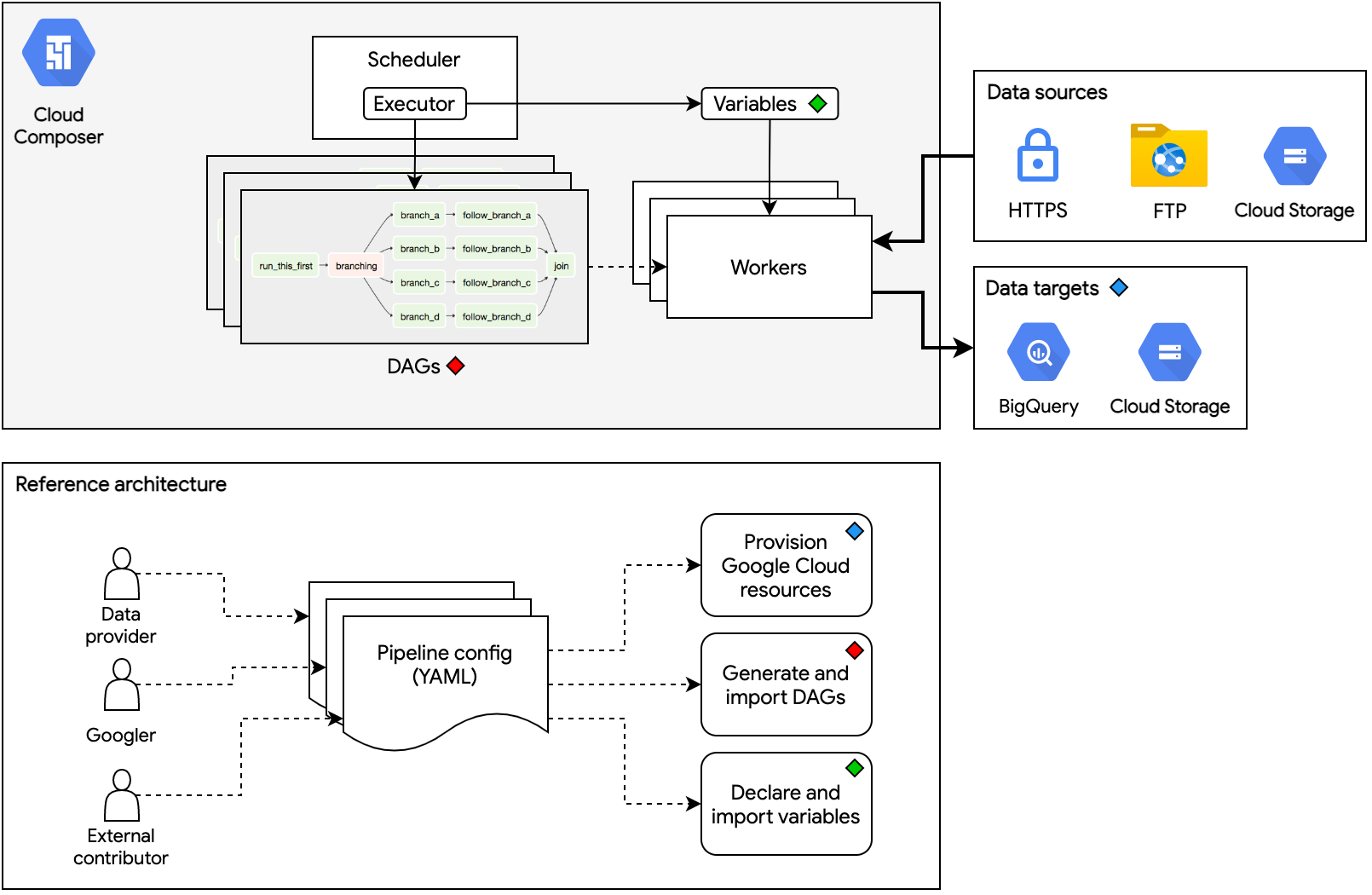
Cloud-native, data pipeline architecture for onboarding public datasets to Datasets for Google Cloud.
- Python
>=3.6.10,<3.9. We currently use3.8. For more info, see the Cloud Composer version list. - Familiarity with Apache Airflow (>=v1.10.15)
- pipenv for creating similar Python environments via
Pipfile.lock - gcloud command-line tool with Google Cloud Platform credentials configured. Instructions can be found here.
- Terraform
>=v0.15.1 - Google Cloud Composer environment running Apache Airflow
>=v1.10.15,<2.0. To create a new Cloud Composer environment, see this guide.
We use Pipenv to make environment setup more deterministic and uniform across different machines.
If you haven't done so, install Pipenv using the instructions found here. Now with Pipenv installed, run the following command:
pipenv install --ignore-pipfile --devThis uses the Pipfile.lock found in the project root and installs all the development dependencies.
Finally, initialize the Airflow database:
pipenv run airflow initdbConfiguring, generating, and deploying data pipelines in a programmatic, standardized, and scalable way is the main purpose of this repository.
Follow the steps below to build a data pipeline for your dataset:
mkdir -p datasets/DATASET/PIPELINE
[example]
datasets/covid19_tracking/national_testing_and_outcomes
where DATASET is the dataset name or category that your pipeline belongs to, and PIPELINE is your pipeline's name.
For examples of pipeline names, see these pipeline folders in the repo.
Use only underscores and alpha-numeric characters for the names.
If you created a new dataset directory above, you need to create a datasets/DATASET/dataset.yaml config file. See this section for the dataset.yaml reference.
Create a datasets/DATASET/PIPELINE/pipeline.yaml config file for your pipeline. See this section for the pipeline.yaml reference.
If you'd like to get started faster, you can inspect config files that already exist in the repository and infer the patterns from there:
- covid19_tracking dataset config
- covid19_tracking/national_testing_and_outcomes pipeline config (simple, only uses built-in Airflow operators)
- covid19_tracking/city_level_cases_and_deaths pipeline config (involves custom data transforms)
Every YAML file supports a resources block. To use this, identify what Google Cloud resources need to be provisioned for your pipelines. Some examples are
- BigQuery datasets and tables to store final, customer-facing data
- GCS bucket to store intermediate, midstream data.
- GCS bucket to store final, downstream, customer-facing data
- Sometimes, for very large datasets, you might need to provision a Dataflow job
Run the following command from the project root:
$ python scripts/generate_terraform.py \
--dataset DATASET_DIR_NAME \
--gcp-project-id GCP_PROJECT_ID \
--region REGION \
--bucket-name-prefix UNIQUE_BUCKET_PREFIX \
[--env] dev \
[--tf-state-bucket] \
[--tf-state-prefix] \
[--tf-apply] \
[--impersonating-acct] IMPERSONATING_SERVICE_ACCTThis generates Terraform files (*.tf) in a _terraform directory inside that dataset. The files contain instrastructure-as-code on which GCP resources need to be actuated for use by the pipelines. If you passed in the --tf-apply parameter, the command will also run terraform apply to actuate those resources.
The --bucket-name-prefix is used to ensure that the buckets created by different environments and contributors are kept unique. This is to satisfy the rule where bucket names must be globally unique across all of GCS. Use hyphenated names (some-prefix-123) instead of snakecase or underscores (some_prefix_123).
The --tf-state-bucket and --tf-state-prefix parameters can be optionally used if one needs to use a remote store for the Terraform state. This will create a backend.tf file that points to the GCS bucket and prefix to use in storing the Terraform state. For more info, see the Terraform docs for using GCS backends.
In addition, the command above creates a "dot" directory in the project root. The directory name is the value you pass to the --env parameter of the command. If no --env argument was passed, the value defaults to dev (which generates the .dev folder).
Consider this "dot" directory as your own dedicated space for prototyping. The files and variables created in that directory will use an isolated environment. All such directories are gitignored.
As a concrete example, the unit tests use a temporary .test directory as their environment.
Run the following command from the project root:
$ python scripts/generate_dag.py \
--dataset DATASET_DIR \
--pipeline PIPELINE_DIR \
[--skip-builds] \
[--env] devThis generates a Python file that represents the DAG (directed acyclic graph) for the pipeline (the dot dir also gets a copy). To standardize DAG files, the resulting Python code is based entirely out of the contents in the pipeline.yaml config file.
Using KubernetesPodOperator requires having a container image available for use. The command above allows this architecture to build and push it to Google Container Registry on your behalf. Follow the steps below to prepare your container image:
-
Create an
_imagesfolder under your dataset folder if it doesn't exist. -
Inside the
_imagesfolder, create another folder and name it after what the image is expected to do, e.g.process_shapefiles,read_cdf_metadata. -
In that subfolder, create a Dockerfile and any scripts you need to process the data. See the
samples/containerfolder for an example. Use the COPY command in yourDockerfileto include your scripts in the image.
The resulting file tree for a dataset that uses two container images may look like
datasets
└── DATASET
├── _images
│ ├── container_a
│ │ ├── Dockerfile
│ │ ├── requirements.txt
│ │ └── script.py
│ └── container_b
│ ├── Dockerfile
│ ├── requirements.txt
│ └── script.py
├── _terraform/
├── PIPELINE_A
├── PIPELINE_B
├── ...
└── dataset.yaml
Docker images will be built and pushed to GCR by default whenever the command above is run. To skip building and pushing images, use the optional --skip-builds flag.
Running the command in the previous step will parse your pipeline config and inform you about the templated variables that need to be set for your pipeline to run.
All variables used by a dataset must have their values set in
[.dev|.test]/datasets/{DATASET}/{DATASET}_variables.json
Airflow variables use JSON dot notation to access the variable's value. For example, if you're using the following variables in your pipeline config:
{{ var.json.shared.composer_bucket }}{{ var.json.parent.nested }}{{ var.json.parent.another_nested }}
then your variables JSON file should look like this
{
"shared": {
"composer_bucket": "us-east4-test-pipelines-abcde1234-bucket"
},
"parent": {
"nested": "some value",
"another_nested": "another value"
}
}
Deploy the DAG and the variables to your own Cloud Composer environment using one of the two commands:
$ python scripts/deploy_dag.py \
--dataset DATASET \
[--pipeline PIPELINE] \
--composer-env CLOUD_COMPOSER_ENVIRONMENT_NAME \
--composer-bucket CLOUD_COMPOSER_BUCKET \
--composer-region CLOUD_COMPOSER_REGION \
--env ENV
The specifying an argument to --pipeline is optional. By default, the script deploys all pipelines under the given --dataset argument.
Run the unit tests from the project root as follows:
$ pipenv run python -m pytest -v
Every dataset and pipeline folder must contain a dataset.yaml and a pipeline.yaml configuration file, respectively:
- For dataset configuration syntax, see
samples/dataset.yamlas a reference. - For pipeline configuration syntax, see
samples/pipeline.yamlas a reference.
-
When running
scripts/generate_terraform.py, the argument--bucket-name-prefixhelps prevent GCS bucket name collisions because bucket names must be globally unique. Use hyphens over underscores for the prefix and make it as unique as possible, and specific to your own environment or use case. -
When naming BigQuery columns, always use
snake_caseand lowercase. -
When specifying BigQuery schemas, be explicit and always include
name,typeandmodefor every column. For column descriptions, derive it from the data source's definitions when available. -
When provisioning resources for pipelines, a good rule-of-thumb is one bucket per dataset, where intermediate data used by various pipelines (under that dataset) are stored in distinct paths under the same bucket. For example:
gs://covid19-tracking-project-intermediate /dev /preprocessed_tests_and_outcomes /preprocessed_vaccinations /staging /national_tests_and_outcomes /state_tests_and_outcomes /state_vaccinations /prod /national_tests_and_outcomes /state_tests_and_outcomes /state_vaccinationsThe "one bucket per dataset" rule prevents us from creating too many buckets for too many purposes. This also helps in discoverability and organization as we scale to thousands of datasets and pipelines.
Quick note: If you can conveniently fit the data in memory, the data transforms are close-to-trivial and are computationally cheap, you may skip having to store mid-stream data. Just apply the transformations in one go, and store the final resulting data to their final destinations.