Datasets for teaching and testing population genetics workflows
remotes::install_github("chollenbeck/popgendata")Load an example simulation dataset in genind format
library(popgendata)
library(pantomime)## Registered S3 method overwritten by 'pegas':
## method from
## print.amova ade4
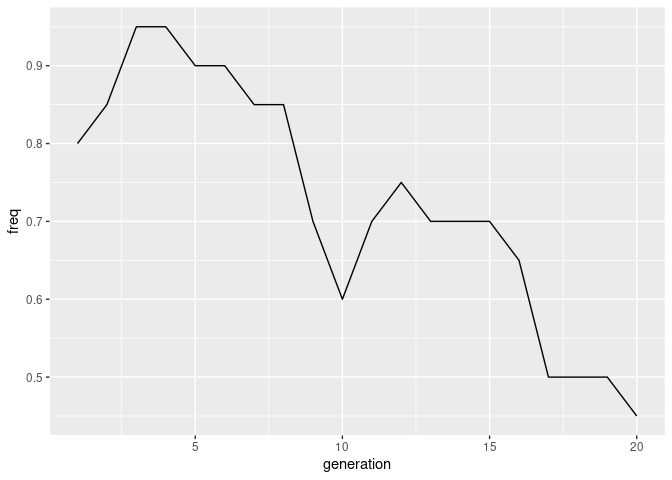
twentygen <- popgendata::twentygenPlot the trajectory of an allele over 20 generations
twentygen %>%
get_allele_freqs() %>% # From pantomime
mutate(generation = as.integer(pop)) %>%
filter(locus == "SNP_1", allele == "G") %>%
ggplot(aes(x = generation, y = freq)) +
geom_line()data("reddrum")
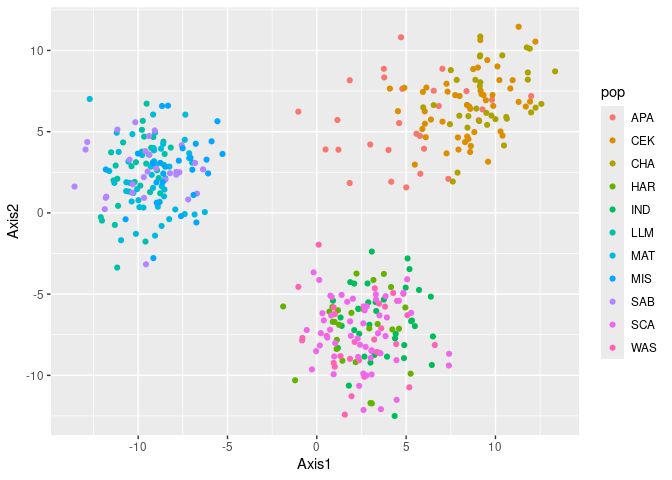
gen <- reddrum$genoRun a quick PCA of the data
pca_tbl <- qpca(gen)
ggplot(pca_tbl, aes(x = Axis1, y = Axis2, col = pop)) +
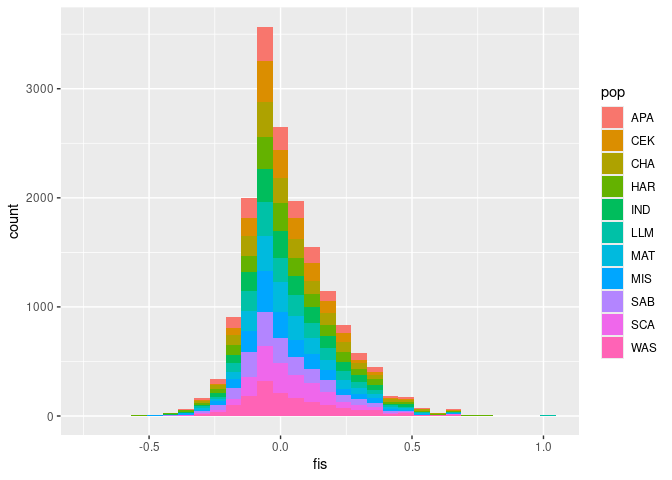
geom_point()Calculate stats by locus and pop and Plot F_{IS}:
stats <- get_locus_stats(gen)
stats## # A tibble: 16,929 × 9
## locus pop n_alleles prop_missing maf ho he fis hwe_pval
## <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <lgl>
## 1 Contig_10008 APA 3 0 0 0.107 0.104 -0.0253 NA
## 2 Contig_10013 APA 3 0.0714 0.0769 0.538 0.469 -0.148 NA
## 3 Contig_10030 APA 2 0 0.214 0.357 0.343 -0.0425 NA
## 4 Contig_1003… APA 2 0 0.179 0.357 0.298 -0.2 NA
## 5 Contig_10045 APA 6 0 0 0.607 0.759 0.200 NA
## 6 Contig_1006… APA 2 0 0.429 0.5 0.499 -0.0027 NA
## 7 Contig_1007… APA 9 0 0 0.714 0.679 -0.0526 NA
## 8 Contig_10089 APA 2 0 0.0536 0.107 0.103 -0.0385 NA
## 9 Contig_1012… APA 2 0.0357 0.315 0.482 0.439 -0.0974 NA
## 10 Contig_10127 APA 2 0 0.0893 0.179 0.165 -0.08 NA
## # ℹ 16,919 more rows
ggplot(stats, aes(x = fis, fill = pop)) +
geom_histogram()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## Warning: Removed 94 rows containing non-finite outside the scale range
## (`stat_bin()`).