This Python client can be used to process in an efficient concurrent manner a set of PDF in a given directory by the GROBID service. Results are written in a given output directory and include the resulting XML TEI representation of the PDF.
You need first to install and start the grobid service, latest stable version, see the documentation. It is assumed that the server will run on the address http://localhost:8070. You can change the server address by editing the file config.json.
This client has been developed and tested with Python 3.5.
Get the github repo:
cd grobid-client-python
There is nothing more to do to start using the python command lines, see the next section.
usage: grobid-client.py [-h] [--input INPUT] [--output OUTPUT]
[--config CONFIG] [--n N] [--generateIDs]
[--consolidate_header] [--consolidate_citations]
[--force]
service
Client for GROBID services
positional arguments:
service one of [processFulltextDocument,
processHeaderDocument, processReferences]
optional arguments:
-h, --help show this help message and exit
--input INPUT path to the directory containing PDF or text to process
--output OUTPUT path to the directory where to put the results (optional)
--config CONFIG path to the config file, default is ./config.json
--n N concurrency for service usage
--generateIDs generate random xml:id to textual XML elements of the
result files
--consolidate_header call GROBID with consolidation of the metadata
extracted from the header
--consolidate_citations
call GROBID with consolidation of the extracted
bibliographical references
--force force re-processing pdf input files when tei output
files already exist
Examples:
python3 grobid-client.py --input ~/tmp/in2 --output ~/tmp/out processFulltextDocument
This command will process all the PDF files present under the input directory recursively (files with extension .pdf only) with the processFulltextDocument service of GROBID, and write the resulting XML TEI files under the output directory, reusing the file name with a different file extension (.tei.xml), using the default 10 concurrent workers.
If --output is omitted, the resulting XML TEI documents will be produced alongside the PDF in the --input directory.
python3 grobid-client.py --input ~/tmp/in2 --output ~/tmp/out --n 20 processHeaderDocument
This command will process all the PDF files present in the input directory (files with extension .pdf only) with the processHeaderDocument service of GROBID, and write the resulting XML TEI files under the output directory, reusing the file name with a different file extension (.tei.xml), using 20 concurrent workers.
By default if an existing .tei.xml file is present in the output directory corresponding to a PDF in the input directory, this PDF will be skipped to avoid reprocessing several times the same PDF. To force the processing of PDF and over-write of existing TEI files, use the parameter --force.
The file test.py gives an example of usage from a another python script.
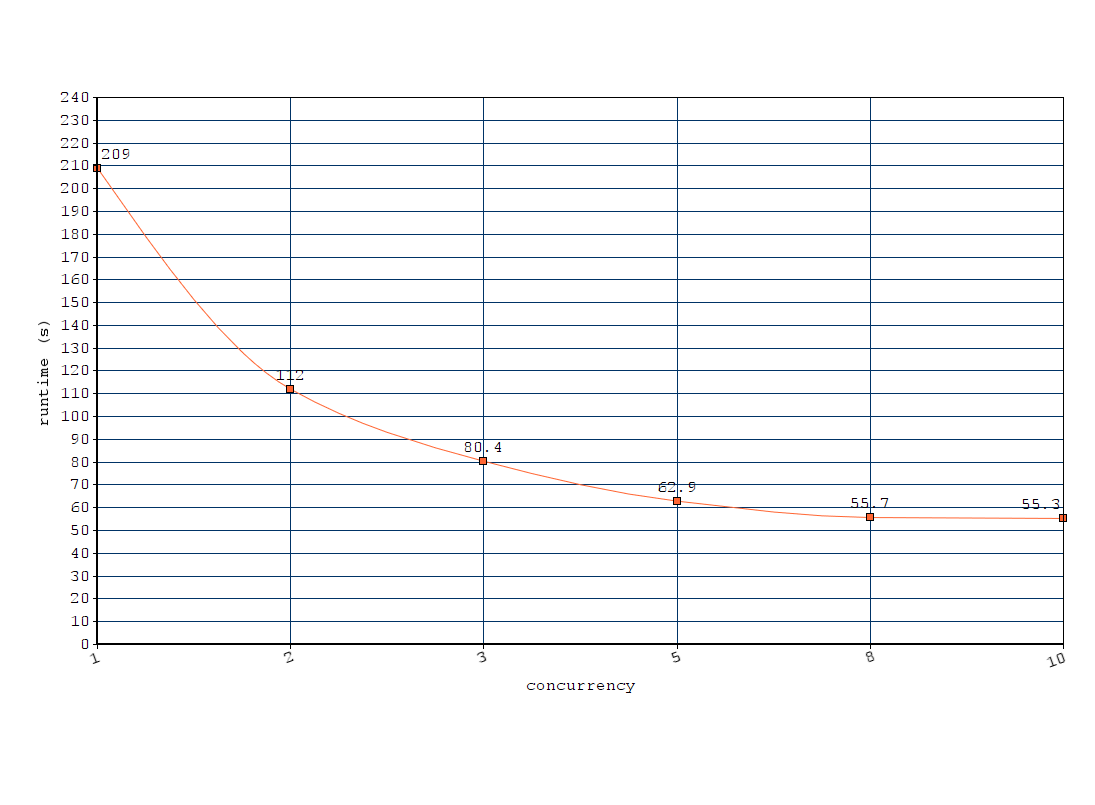
Full text processing of 136 PDF (total 3443 pages, in average 25 pages per PDF) on Intel Core i7-4790K CPU 4.00GHz, 4 cores (8 threads), 16GB memory, n being the concurrency parameter:
| n | runtime (s) | s/PDF | PDF/s |
|---|---|---|---|
| 1 | 209.0 | 1.54 | 0.65 |
| 2 | 112.0 | 0.82 | 1.21 |
| 3 | 80.4 | 0.59 | 1.69 |
| 5 | 62.9 | 0.46 | 2.16 |
| 8 | 55.7 | 0.41 | 2.44 |
| 10 | 55.3 | 0.40 | 2.45 |
As complementary info, GROBID processing of header of the 136 PDF and with n=10 takes 3.74 s (15 times faster than the complete full text processing because only the two first pages of the PDF are considered), 36 PDF/s. In similar conditions, extraction and structuring of bibliographical references takes 26.9 s (5.1 PDF/s).
Benchmarking with many more files (e.g. million ISTEX PDF). Also implement existing GROBID services for text input (date, name, affiliation/address, raw bibliographical references, etc.). Better support for parameters (including elements where to put coordinates).
Distributed under Apache 2.0 license.
Main author and contact: Patrice Lopez (patrice.lopez@science-miner.com)