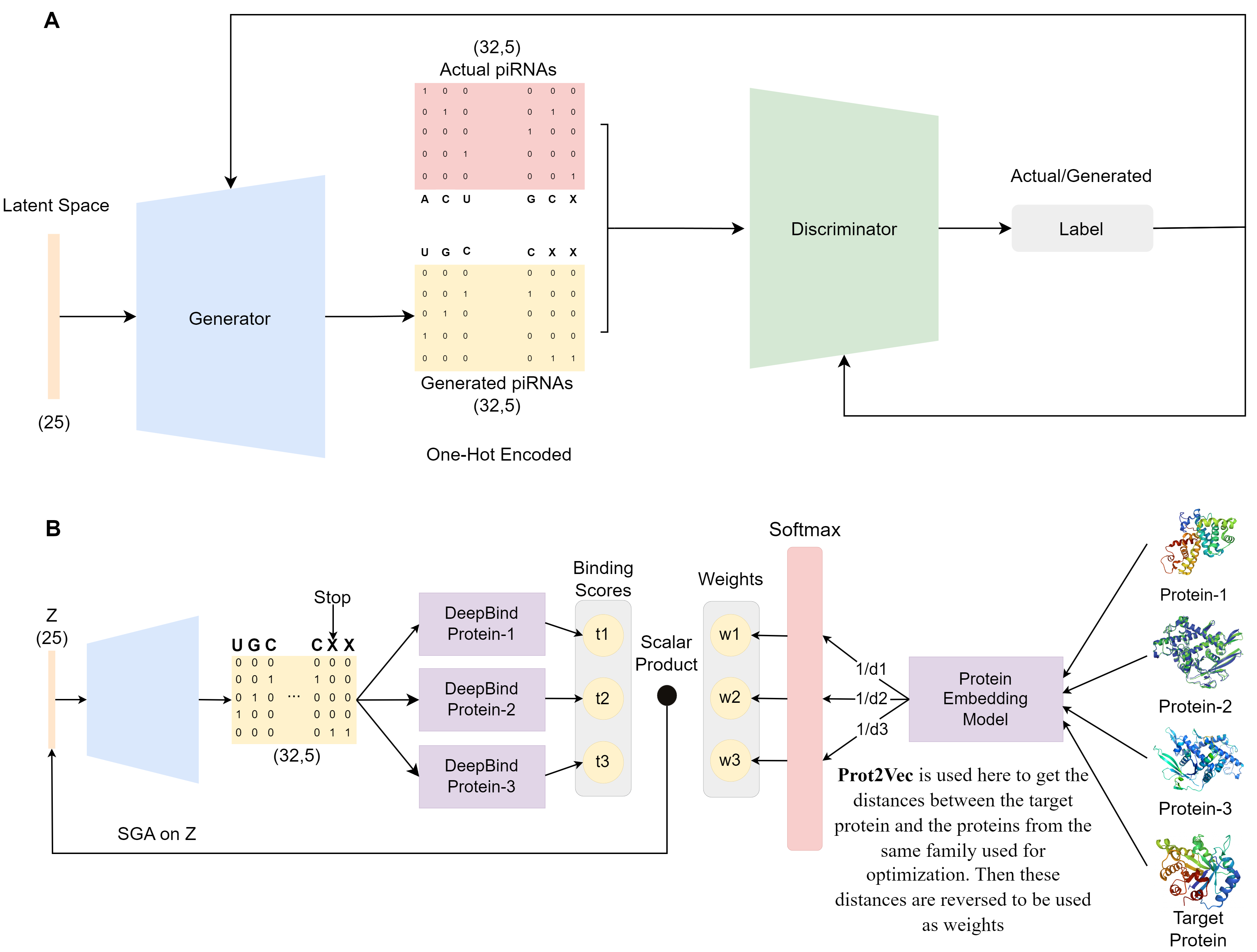
RNAGEN is a is a deep learning based model for novel piRNA generation and optimziation. We use the WGAN-GP architecture for the generative model, and the DeepBind models for optimizing binding of the generated piRNA sequences to the target protein. To find the closest relatives of a target protein to be used in optimization, we use the Prot2Vec model.
Diagram of the generative model and the optimization procedure
Furkan Ozden, Sina Barazandeh, Dogus Akboga, Urartu Ozgur Safak Seker, A. Ercument Cicek
[secondauthorname].[secondauthorsurname]@bilkent.edu.tr
[firstcorrespondingauthorfirstname]@bilkent.edu.tr
[secondcorrespondingauthorsurname]@cs.bilkent.edu.tr
Warning: Please note that the RNAGEN model is completely free for academic usage. However it is licenced for commercial usage. Please first refer to the License section for more info.
- RNAGEN is easy to use and does not require installation. The scripts can be used if the requirements are installed.
Note: The implementation is using Tensorflow 1.15, but the provided code uses Tensorflow 2 for easier installation and use. Tensorflow 1 is available in Tensorflow 2 using the tf.compat.v1 module.
For easy requirement handling, you can use RNAGEN.yml files to initialize conda environment with requirements installed:
$ conda env create --name rnagen -f RNAGEN.yml
$ conda activate rnagenNote that the provided environment yml file is for Linux systems. For MacOS users, the corresponding versions of the packages might need to be changed.
- RNAGEN components are trained using GPUs and GPUs are used for the project. However, depending on the type of Tensorflow Tensorflow the model can run on both GPU and CPU. The run time on CPU is considerably longer compared to GPU.
- ./analysis/binding_plot.py : plots the binding score plots on the manuscript
- ./analysis/gan_validation.py : plots gan validation plots on the manuscript
- ./analysis/generate.py : generates the ./analysis/generated.txt sequences using the trained model
- ./analysis/gen_ham_dist.npy : saved array of distances between the generated and the natural set of sequences (regenerated if removed)
- ./analysis/rand_ham_dist.npy : saved array of distances between the random and the natural set of sequences (regenerated if removed)
- ./analysis/real_ham_dist.npy : saved array of non-zero distances between the set of natural sequences and itself (regenerated if removed)
- ./analysis/generated.txt : generated using the trained generator
- ./analysis/piRNAs.fa : set of natural sequences extracted from ./data/DASHR2_GEO_hg38_sequenceTable_export.csv
- ./data/model/ : the trained generator model
- ./data/DASHR2_GEO_hg38_sequenceTable_export.csv : the original dataset used for training and validation
- ./data/protVec_100d_3grams.csv : taken from Prot2Vec, used for calculating distances between proteins
- ./deepbind_models : taken directly from the deepbind directory
- ./figures/ : directory for the generated figures
- ./input/opt_input.csv : input sample for the ./optimize.py script
- ./input/proteins.csv : input sample for the ./distance_proteins.py script
- ./lib/ : utility files for the implementation
- ./output/ : output of the ./optimize.py are saved here (SOX2, SOX4 already exist).
- ./distance_proteins.py : calculates distances between sequences for the input csv file
- ./optimize.py : optimizes generated piRNAs based on the given input csv file
- ./train_rnagen.py : trains the RNAGEN generator using the natural sequences
Important notice: Please call the train_rnagen.py script from the root directory. The optimization can be performed using the optimize.py script. To analyze the generated seqeunces use the ./analysis/gan_validation.py script. This script will generate plots that are used in the manuscript to validate the GAN model.
To plot the optimization and initial binding score plots, you need to first run the optimization, and then use the ./analysis/binding_plot.py by giving the required arguments. These arguments will be used to find the optimization output directory for plotting.
$ python ./train_rnagen.py- Minimum length of the piRNAs used for training the model. Default: 26.
- Maximum length of the piRNAs used for training the model. Default: 32.
- The path to file including the piRNA samples. The default path used is './data/DASHR2_GEO_hg38_sequenceTable_export.csv'.
- The learning rate of the Adam optimizer used to optimize the model parameters. The default value is 1e-5. If 4 is provided, the learning rate will be 1e-4.
- The batch size used to train the model, the default value is 64
- The dimension of the input noise, with the default value of 40.
- The ID of the GPU that will be used for training. The default setting is used otherwise.
The output of the training will be in './logs/gan_test/{timestamp}/'.
$ python ./distance_proteins.py- The path to a text file including protein names and sequences. The default path is './input/proteins.csv' and by editing that file, the script will use those sequences and names.
The file includes at least 2 proteins. The first one is the target protein, ones are the protines for which we want to calculate the distance.
The output will be shown on the terminal. It will include the protein names and their distances to the target protein in the Prot2Vec space.
$ python ./optimize.py- The path to a text file including paths of the deepbind models of the relative proteins. The default path is './input/opt_input.csv' and by editing that file, the optimization will use those files and proteins.
The file includes 5 proteins. The first one is the target protein, the next three are the chosen relative proteins for optimization, and the last one is optional test protein in case you want to test the results on a different protein (disabled for now).
- The path to the tarined generator model. Default value is './data/model/trained_gan.ckpt.meta'.
- The learning rate of the Adam optimizer used to optimize the model parameters. The default value is 3e-5.
- The number of iterations the optimization is performed. The default value is 3,000 iterations.
- The id of the gpu used for optimization. The default value will be used otherwise.
- The optimiztation outputs will be put in './output/'. The file names are descriptive.
$ python ./analysis/generate.py- It will output the './analysis/generated.txt' file.
$ python ./analysis/gan_validation.py- It will output plots in the './figures/' folder
$ python ./analysis/binding_plots.py- The name of the first protein (SOX4 as default).
- The name of the first protein (SOX2 as default).
- The number of optimization iterations for the first protein (3000 as default).
- The number of optimization iterations for the second protein (3000 as default).
- It will output plots in the './figures/' folder
Usage of RNAGEN is very simple. You need to install conda to install the specific environment and run the scripts.
-
This project uses conda package management software to create virtual environment and facilitate reproducability.
-
For Linux users:
-
Please take a look at the Anaconda repo archive page, and select an appropriate version that you'd like to install.
-
Replace this
Anaconda3-version.num-Linux-x86_64.shwith your choice
$ wget -c https://repo.continuum.io/archive/Anaconda3-vers.num-Linux-x86_64.sh
$ bash Anaconda3-version.num-Linux-x86_64.sh- It is important to set up the conda environment which includes the necessary dependencies.
- Please run the following lines to create and activate the environment:
$ conda env create --name rnagen -f RNAGEN.yml
$ conda activate rnagenThe study is available as a preprint on biorixv. https://www.biorxiv.org/content/10.1101/2023.07.11.548246v1
- CC BY-NC-SA 2.0
- Copyright 2023 © RNAGEN.
- For commercial usage, please contact.