My R toolkit for organizing analysis projects, cleaning data for machine learning, parallelizing code for multiple cores or in a SLURM cluster, and extended functionality for SuperLearner and TMLE. Some of the SuperLearner functions may eventually be migrated into the SuperLearner package.
Install the latest release from CRAN:
install.packages("ck37r") Install the development version from github (recommended):
# install.packages("remotes")
remotes::install_github("ck37/ck37r")- Project Utilities
import_csvs- import all CSV files in a given directory.load_all_code- source() all R files in a given directory.load_packages- load a list of packages; for the ones that fail it can attempt to install them automatically from CRAN, then load them again.
- Machine Learning
categoricals_to_factors- convert numeric categoricals into factors.factors_to_indicators- convert all factors in a dataframe to series of indicators (one-hot encoding).impute_missing_values- impute missing values in a dataframe (median for numerics and mode for factors, GLRM, or k-nearest neighbors), add missingness indicators.missingness_indicators- return a matrix of missingness indicators for a dataframe, (optionally) omitting any constant or collinear columns.rf_count_terminal_nodes- count the number of terminal nodes in each tree in a random forest. That information can then be used to grid-search the maximum number of nodes allowed in a Random Forest (along with mtry).standardize- standardize a dataset (center, scale), optionally omitting certain variables.vim_corr- rudimentary variable importance based on correlation with an outcome.
- Parallelization
parallelize- starts a multicore or multinode parallel cluster. Automatically detects parallel nodes in a SLURM environment, which makes code work seemlessly on a laptop or a cluster.stop_cluster- stops a cluster started byparallelize().
- SuperLearner
auc_table- table of cross-validated AUCs for each learner in an ensemble, including SE, CI, and p-value. Supports SuperLearner and CV.SuperLearner objects.gen_superlearner- create a SuperLearner and CV.SuperLearner function setup to transparently use a certain parallelization configuration.cvsl_weights- table of the meta-weight distribution for each learner in a CV.SuperLearner analysis.cvsl_auc- cross-validated AUC for a CV.SuperLearner analysis.plot_roc- ROC plot with AUC and CI for a SuperLearner or CV.SuperLearner object.plot.SuperLearner- plot risk estimates and CIs for a SuperLearner, similar to CV.Superlearner except without SL or Discrete SL.prauc_table- table of cross-validated PR-AUCs for each learner in an ensemble, including SE and CI. Supports SuperLearner and CV.SuperLearner objects.sl_stderr- calculate standard error for each learner’s risk in SL.SL.h2o_auto()- wrapper for h2o’s automatic machine learning system, to be added to SuperLearner.SL.bartMachine2()- wrapper for bartMachine, to be added to SuperLearner.
- TMLE
tmle_parallel- allows the SuperLearner estimation in TMLE to be customized, esp. to support parallel estimation via mcSuperLearner and snowSuperLearner.setup_parallel_tmle- helper function to start a cluster and setup SuperLearner and tmle_parallel to use the created cluster.
- h2o
h2o_init_multinode()- function to start an h2o cluster on multiple nodes from within R, intended for use on SLURM or other multi-node clusters.
# Load a test dataset.
# TODO: need to switch to a different dataset.
data(PimaIndiansDiabetes2, package = "mlbench")
# Check for missing values.
colSums(is.na(PimaIndiansDiabetes2))
#> pregnant glucose pressure triceps insulin mass pedigree age
#> 0 5 35 227 374 11 0 0
#> diabetes
#> 0
# Impute missing data and add missingness indicators.
# Don't impute the outcome though.
result = impute_missing_values(PimaIndiansDiabetes2, skip_vars = "diabetes")
# Confirm we have no missing data.
colSums(is.na(result$data))
#> pregnant glucose pressure triceps insulin
#> 0 0 0 0 0
#> mass pedigree age diabetes miss_glucose
#> 0 0 0 0 0
#> miss_pressure miss_triceps miss_insulin miss_mass
#> 0 0 0 0We are using default hyperparameters here, but it would be best to optimize the hyperparameters.
#############
# Generalized low-rank model imputation via h2o.
result2 = impute_missing_values(PimaIndiansDiabetes2, type = "glrm", skip_vars = "diabetes")
# Confirm we have no missing data.
colSums(is.na(result2$data))
#> pregnant glucose pressure triceps insulin
#> 0 0 0 0 0
#> mass pedigree age diabetes miss_glucose
#> 0 0 0 0 0
#> miss_pressure miss_triceps miss_insulin miss_mass
#> 0 0 0 0This loads a vector of packages, automatically installing any packages that aren’t already installed.
# Load these 4 packages and install them if necessary.
load_packages(c("MASS", "SuperLearner", "tmle", "doParallel"), auto_install = TRUE)
#> Loaded gam 1.20
#> Super Learner
#> Version: 2.0-28
#> Package created on 2021-05-04
#> Loaded glmnet 4.1-3
#> Welcome to the tmle package, version 1.5.0-1.1
#>
#> Major changes since v1.3.x. Use tmleNews() to see details on changes and bug fixesWe estimate one standard Random Forest first and examine how many terminal nodes are in each decision tree. We take the maximum of that as the most data-adaptive Random Forest in terms of decision tree size, then compare to Random Forests in which they are restricted to have smaller decision trees. This allows the SuperLearner to explore under vs. over-fitting for a Random Forest. See Segal (2004) and Segal & Xiao (2011) for details on overfitting in Random Forests.
library(SuperLearner)
library(ck37r)
data(Boston, package = "MASS")
set.seed(1)
(sl = SuperLearner(Boston$medv, subset(Boston, select = -medv), family = gaussian(),
cvControl = list(V = 3L),
SL.library = c("SL.mean", "SL.glm", "SL.randomForest")))
#> Loading required namespace: randomForest
#>
#> Call:
#> SuperLearner(Y = Boston$medv, X = subset(Boston, select = -medv), family = gaussian(),
#> SL.library = c("SL.mean", "SL.glm", "SL.randomForest"), cvControl = list(V = 3L))
#>
#>
#>
#> Risk Coef
#> SL.mean_All 85.22648 0.000000
#> SL.glm_All 25.31448 0.063065
#> SL.randomForest_All 12.89105 0.936935
summary(rf_count_terminal_nodes(sl$fitLibrary$SL.randomForest_All$object))
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 133.0 163.0 167.0 166.6 171.0 187.0
(max_terminal_nodes = max(rf_count_terminal_nodes(sl$fitLibrary$SL.randomForest_All$object)))
#> [1] 187
# Now run create.Learner() based on that maximum.
# It is often handy to convert to log scale of a hyperparameter before testing a ~linear grid.
# NOTE: -0.7 ~ log(0.5) which is the multiplier that yields sqrt(max)
(maxnode_seq = unique(round(exp(log(max_terminal_nodes) * exp(c(-0.6, -0.35, -0.15, 0))))))
#> [1] 18 40 90 187
rf = create.Learner("SL.randomForest", detailed_names = TRUE,
name_prefix = "rf",
params = list(ntree = 100L), # fewer trees for testing speed only.
tune = list(maxnodes = maxnode_seq))
# We see that an RF with simpler decision trees performs better than the default.
(sl = SuperLearner(Boston$medv, subset(Boston, select = -medv), family = gaussian(),
cvControl = list(V = 3L),
SL.library = c("SL.mean", "SL.glm", rf$names)))
#>
#> Call:
#> SuperLearner(Y = Boston$medv, X = subset(Boston, select = -medv), family = gaussian(),
#> SL.library = c("SL.mean", "SL.glm", rf$names), cvControl = list(V = 3L))
#>
#>
#> Risk Coef
#> SL.mean_All 84.50244 0.0000000
#> SL.glm_All 24.49011 0.0000000
#> rf_18_All 13.47510 0.0000000
#> rf_40_All 11.82894 0.0000000
#> rf_90_All 11.09016 0.5311252
#> rf_187_All 11.08278 0.4688748library(ck37r)
library(tmle)
# Use multiple cores as available.
ck37r::setup_parallel_tmle()
# Basic SL library.
sl_lib = c("SL.mean", "SL.rpart", "SL.glmnet")
# Set a parallel-compatible seed so cross-validation folds are deterministic.
set.seed(1, "L'Ecuyer-CMRG")
# Just an example -- we haven't defined A or W in this code.
result = run_tmle(Y = Y, A = A, W = W, family = "binomial",
g.SL.library = sl_lib, Q.SL.library = sl_lib)This will return an AUC table for all learners. It does not include Discrete SL or SuperLearner as those require CV.SuperLearner.
library(SuperLearner)
library(ck37r)
data(Boston, package = "MASS")
set.seed(1)
y = as.numeric(Boston$medv > 23)
sl = SuperLearner(Y = y,
X = subset(Boston, select = -medv),
family = binomial(),
cvControl = list(V = 2L, stratifyCV = TRUE),
SL.library = c("SL.mean", "SL.lm", "SL.glm"))
auc_table(sl, y = y)
#> learner auc se ci_lower ci_upper p-value
#> 1 SL.mean_All 0.5000000 0.04590129 0.4100351 0.5899649 2.560257e-21
#> 2 SL.lm_All 0.9276482 0.01241868 0.9033081 0.9519884 3.697143e-01
#> 3 SL.glm_All 0.9317788 0.01212514 0.9080140 0.9555437 5.000000e-01This is similar to CV.SuperLearner’s plot except SuperLearner cannot estimate risk for the Discrete SL and SuperLearner, so those must be omitted here.
library(SuperLearner)
library(ck37r)
data(Boston, package = "MASS")
set.seed(1)
(sl = SuperLearner(Boston$medv, subset(Boston, select = -medv),
family = gaussian(),
cvControl = list(V = 2),
SL.library = c("SL.mean", "SL.lm")))
#>
#> Call:
#> SuperLearner(Y = Boston$medv, X = subset(Boston, select = -medv), family = gaussian(),
#> SL.library = c("SL.mean", "SL.lm"), cvControl = list(V = 2))
#>
#>
#> Risk Coef
#> SL.mean_All 84.68179 0.03641976
#> SL.lm_All 24.95053 0.96358024
plot(sl, y = Boston$medv)library(SuperLearner)
library(ck37r)
data(Boston, package = "MASS")
y = as.numeric(Boston$medv > 23)
set.seed(1)
(sl = SuperLearner(Y = y,
X = subset(Boston, select = -medv),
family = binomial(),
cvControl = list(V = 2L, stratifyCV = TRUE),
SL.library = c("SL.mean", "SL.lm", "SL.glm")))
#>
#> Call:
#> SuperLearner(Y = y, X = subset(Boston, select = -medv), family = binomial(),
#> SL.library = c("SL.mean", "SL.lm", "SL.glm"), cvControl = list(V = 2L,
#> stratifyCV = TRUE))
#>
#>
#> Risk Coef
#> SL.mean_All 0.2344983 0.0000000
#> SL.lm_All 0.1107146 0.1730418
#> SL.glm_All 0.0976933 0.8269582
plot_roc(sl, y = y)Reports on the precision-recall AUC for each learner, and includes the estimated standard error and 95% confidence interval.
library(SuperLearner)
library(ck37r)
data(Boston, package = "MASS")
y = as.numeric(Boston$medv > 23)
set.seed(1)
sl = SuperLearner(Y = y,
X = subset(Boston, select = -medv),
family = binomial(),
cvControl = list(V = 2L, stratifyCV = TRUE),
SL.library = c("SL.mean", "SL.lm", "SL.glm"))
prauc_table(sl, y = y)
#> learner prauc stderr ci_lower ci_upper
#> 1 SL.mean_All 0.3754941 0.04968302 0.2841487 0.4766516
#> 2 SL.lm_All 0.9008753 0.03058504 0.8205770 0.9475348
#> 3 SL.glm_All 0.9041609 0.03005690 0.8221817 0.9506155This will return the AUC inference for the CV.SuperLearner.
library(SuperLearner)
library(ck37r)
data(Boston, package = "MASS")
set.seed(1)
cvsl = CV.SuperLearner(Y = as.numeric(Boston$medv > 23),
X = subset(Boston, select = -medv),
family = binomial(),
cvControl = list(V = 2L, stratifyCV = TRUE),
SL.library = c("SL.mean", "SL.lm", "SL.glm"))
cvsl_auc(cvsl)
#> $cvAUC
#> [1] 0.9339107
#>
#> $se
#> [1] 0.01161126
#>
#> $ci
#> [1] 0.9111531 0.9566684
#>
#> $confidence
#> [1] 0.95This will return an AUC table for all learners, plus DiscreteSL and the SuperLearner.
library(SuperLearner)
library(ck37r)
data(Boston, package = "MASS")
set.seed(1)
cvsl = CV.SuperLearner(Y = as.numeric(Boston$medv > 23),
X = subset(Boston, select = -medv),
family = binomial(),
cvControl = list(V = 2, stratifyCV = TRUE),
SL.library = c("SL.mean", "SL.lm", "SL.glm"))
auc_table(cvsl)
#> auc se ci_lower ci_upper p-value
#> SL.mean_All 0.5000000 0.04590129 0.4100351 0.5899649 1.644292e-21
#> SL.lm_All 0.9276482 0.01241868 0.9033081 0.9519884 3.070323e-01
#> SL.glm_All 0.9317788 0.01212514 0.9080140 0.9555437 4.302154e-01
#> DiscreteSL 0.9317788 0.01212514 0.9080140 0.9555437 4.302154e-01
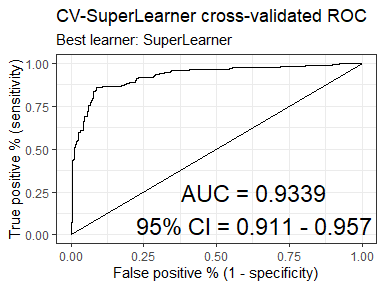
#> SuperLearner 0.9339107 0.01161126 0.9111531 0.9566684 5.000000e-01library(SuperLearner)
library(ck37r)
data(Boston, package = "MASS")
set.seed(1)
cvsl = CV.SuperLearner(Y = as.numeric(Boston$medv > 23),
X = subset(Boston, select = -medv),
family = binomial(),
cvControl = list(V = 2L, stratifyCV = TRUE),
SL.library = c("SL.mean", "SL.lm", "SL.glm"))
plot_roc(cvsl)Returns summary statistics (mean, sd, min, max) on the distribution of the weights assigned to each learner across SuperLearner ensembles. This makes it easier to understand the stochastic nature of the SL learner weights and to see how often certain learners are used.
library(SuperLearner)
library(ck37r)
data(Boston, package = "MASS")
set.seed(1)
cvsl = CV.SuperLearner(Y = as.numeric(Boston$medv > 23),
X = subset(Boston, select = -medv),
family = binomial(),
cvControl = list(V = 2L, stratifyCV = TRUE),
SL.library = c("SL.mean", "SL.lm", "SL.glm"))
cvsl_weights(cvsl)
#> # Learner Mean SD Min Max
#> 1 1 glm 0.82899 0.10478 0.75490 0.90308
#> 2 2 lm 0.14422 0.14266 0.04335 0.24510
#> 3 3 mean 0.02679 0.03788 0.00000 0.05357Reports on the precision-recall AUC for each learner, as well as the SuperLearner and Discrete Superlearner. Includes the estimated standard error and 95% confidence interval.
library(SuperLearner)
library(ck37r)
data(Boston, package = "MASS")
set.seed(1)
cvsl = CV.SuperLearner(Y = as.numeric(Boston$medv > 23),
X = subset(Boston, select = -medv),
family = binomial(),
cvControl = list(V = 2L, stratifyCV = TRUE),
SL.library = c("SL.mean", "SL.lm", "SL.glm"))
prauc_table(cvsl)
#> prauc stderr ci_lower ci_upper
#> SL.mean_All 0.3754941 0.04968302 0.2841487 0.4766516
#> SL.lm_All 0.9008753 0.03058504 0.8205770 0.9475348
#> SL.glm_All 0.9041609 0.03005690 0.8221817 0.9506155
#> DiscreteSL 0.9041609 0.03005690 0.8221817 0.9506155
#> SuperLearner 0.9102754 0.02922821 0.8308581 0.9544482More examples to be added.
The ROC plot is inspired by similar base plot functionality created by Alan Hubbard.
Breiman, L. (2001). Random forests. Machine learning, 45(1), 5-32.
Dudoit, S., & van der Laan, M. J. (2005). Asymptotics of cross-validated risk estimation in estimator selection and performance assessment. Statistical Methodology, 2(2), 131-154.
LeDell, E., Petersen, M., & van der Laan, M. (2015). Computationally efficient confidence intervals for cross-validated area under the ROC curve estimates. Electronic journal of statistics, 9(1), 1583.
Polley EC, van der Laan MJ (2010) Super Learner in Prediction. U.C. Berkeley Division of Biostatistics Working Paper Series. Paper 226. http://biostats.bepress.com/ucbbiostat/paper266/
Segal, M. R. (2004). Machine learning benchmarks and random forest regression.
Segal, M., & Xiao, Y. (2011). Multivariate random forests. Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery, 1(1), 80-87.
Sing, T., Sander, O., Beerenwinkel, N., & Lengauer, T. (2005). ROCR: visualizing classifier performance in R. Bioinformatics, 21(20), 3940-3941.
van der Laan, M. J., Polley, E. C. and Hubbard, A. E. (2007) Super Learner. Statistical Applications of Genetics and Molecular Biology, 6, article 25. http://www.degruyter.com/view/j/sagmb.2007.6.issue-1/sagmb.2007.6.1.1309/sagmb.2007.6.1.1309.xml
van der Laan, M. J., & Rose, S. (2011). Targeted learning: causal inference for observational and experimental data. Springer Science & Business Media.
van der Laan, M. J., & Rubin, D. (2006). Targeted Maximum Likelihood Learning. The International Journal of Biostatistics, 2(1), 1-38.