▄████████ ▄█ ▄████████ ▄▄▄▄███▄▄▄▄ ▄██ ▄ ███ ▄██████▄ ▄████████
███ ███ ███ ███ ███ ▄██▀▀▀███▀▀▀██▄ ███ ██▄ ▀█████████▄ ███ ███ ███ ███
███ █▀ ███ ███ ███ ███ ███ ███ ███▄▄▄███ ▀███▀▀██ ███ ███ ███ █▀
███ ███ ███ ███ ███ ███ ███ ▀▀▀▀▀▀███ ███ ▀ ███ ███ ▄███▄▄▄
███ ███ ▀███████████ ███ ███ ███ ▄██ ███ ███ ███ ███ ▀▀███▀▀▀
███ █▄ ███ ███ ███ ███ ███ ███ ███ ███ ███ ███ ███ ███ █▄
███ ███ ███▌ ▄ ███ ███ ███ ███ ███ ███ ███ ███ ███ ███ ███ ███
████████▀ █████▄▄██ ███ █▀ ▀█ ███ █▀ ▀█████▀ ▄████▀ ▀██████▀ ██████████
▀ This classifier will predict the likelyhood of someone dying from taking the covid vaccination. The dataset was collected from the Vaccine Adverse Event Reporting System (VAERS), which is an arm of the U.S. Department of Health & Human Services (HHS).
If you are interested in the actual raw data, the datasets can be downloaded from the VAERS Data Sets: https://vaers.hhs.gov/data/datasets.html.
These are the two datasets that were used:
- 2022VAERSDATA.csv
- 2022VAERSVAX.csv
The fastest way for you to run this model is to simply get the docker image clamytoe/covid_risk_classifier and run it locally.
docker pull clamytoe/covid_risk_classifier
docker run -it --rm -p 3000:3000 clamytoe/covid_risk_classifier serve --production
You can use the API by simply opening a browser to http://localhost:3000
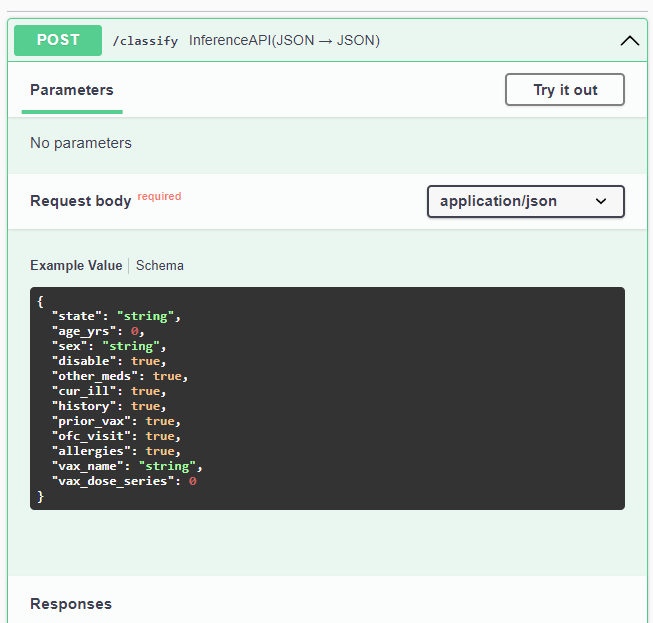
If you scroll down a bit until you get to the Service APIs section and click on the POST drop down.
Once you've clicked on it to expand it, you will see a Try it out button.
Clicking on it will expose a text area with a schema of a valid input.
Enter your own values to try it out. Due take note that at the top of the page, a description of each value is provided along with acceptable values.
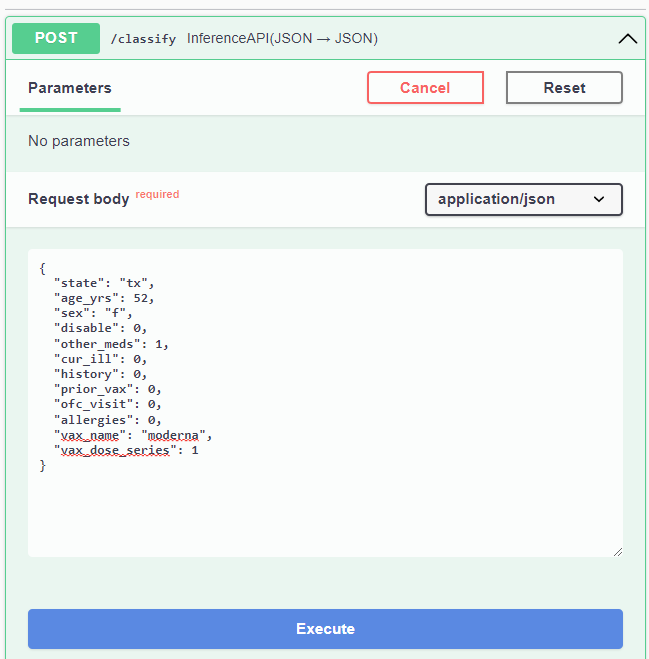
Here is a sample for you to try:
{
"state": "tx",
"age_yrs": 52,
"sex": "f",
"disable": 0,
"other_meds": 1,
"cur_ill": 0,
"history": 0,
"prior_vax": 0,
"ofc_visit": 0,
"allergies": 0,
"vax_name": "moderna",
"vax_dose_series": 1
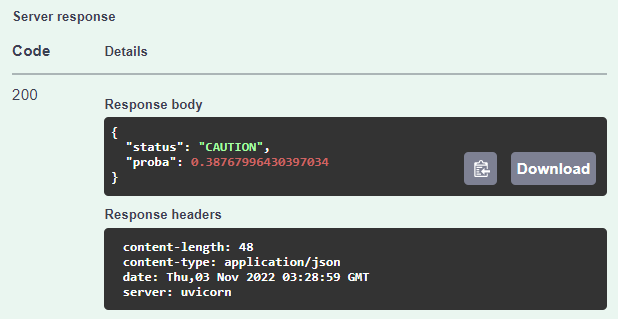
}Once you've entered your values, just click on the Execute button and scroll down a bit until you get to the Server response section.
In this case, we see that the user should be cautious if planning to take it.
If entering each patient's data takes too long, you can do something like what I'm doing with my predict.py script.
predict.py:
import json
import requests
from service import CovidRisk
patients: list[CovidRisk] = [
{
"state": "mi",
"age_yrs": 85,
"sex": "f",
"disable": 0,
"other_meds": 0,
"cur_ill": 0,
"history": 0,
"prior_vax": 0,
"ofc_visit": 0,
"allergies": 0,
"vax_name": "pfizer\\biontech",
"vax_dose_series": 2,
},
...
{
"state": "tx",
"age_yrs": 52,
"sex": "f",
"disable": 0,
"other_meds": 1,
"cur_ill": 0,
"history": 0,
"prior_vax": 0,
"ofc_visit": 0,
"allergies": 0,
"vax_name": "moderna",
"vax_dose_series": 1,
},
]
def test_service(data: List[CovidRisk]):
for patient in data:
patient = json.dumps(patient)
response = requests.post(
"http://localhost:3000/classify",
headers={"content-type": "application/json"},
data=patient,
).json()
print(response["status"], response["proba"])
if __name__ == "__main__":
test_service(patients)I've enabled logging for the service. In order to be able to keep the logs you will have to create a local mount point or a docker volume. That's beyong the scope of this project, so I will leave that up to you to implement on your own.
- If you run the service locally, it is created in your project's root directory as
covid_risk_classifier.log. - If you run it from a docker image, it is created in
/home/bentoml/bento/srcwith the same name.
sample log entries:
cat covid_risk_classifier.log
...
2022-11-04T23:01:51.342665+0000 INFO Processing: state='tx' age_yrs=53 sex='f' disable=False other_meds=False cur_ill=False history=False prior_vax=False ofc_visit=False allergies=False vax_name='pfizer\\biontech' vax_dose_series=1
2022-11-04T23:01:51.347936+0000 INFO Probability: 0.19510182738304138
2022-11-04T23:01:51.348160+0000 INFO Prediction: {'status': 'SAFE', 'proba': 0.19510183}
2022-11-04T23:01:51.351920+0000 INFO Processing: state='ca' age_yrs=3 sex='m' disable=False other_meds=False cur_ill=False history=False prior_vax=False ofc_visit=False allergies=False vax_name='moderna' vax_dose_series=1
2022-11-04T23:01:51.357594+0000 INFO Probability: 0.003593571949750185
2022-11-04T23:01:51.357773+0000 INFO Prediction: {'status': 'SAFE', 'proba': 0.003593572}
2022-11-04T23:01:51.361099+0000 INFO Processing: state='tx' age_yrs=52 sex='f' disable=False other_meds=True cur_ill=False history=False prior_vax=False ofc_visit=False allergies=False vax_name='moderna' vax_dose_series=1
2022-11-04T23:01:51.366562+0000 INFO Probability: 0.13526801764965057
2022-11-04T23:01:51.366764+0000 INFO Prediction: {'status': 'SAFE', 'proba': 0.13526802}
If you want to dig in more and take a look at what it takes to create your own classifier, then a look at the HOW-TO.md file.