Enzyme is a plugin that performs automatic differentiation (AD) of statically analyzable LLVM and MLIR.
Enzyme can be used by calling __enzyme_autodiff on a function to be differentiated as shown below.
Running the Enzyme transformation pass then replaces the call to __enzyme_autodiff with the gradient of its first argument.
double foo(double);
double grad_foo(double x) {
return __enzyme_autodiff(foo, x);
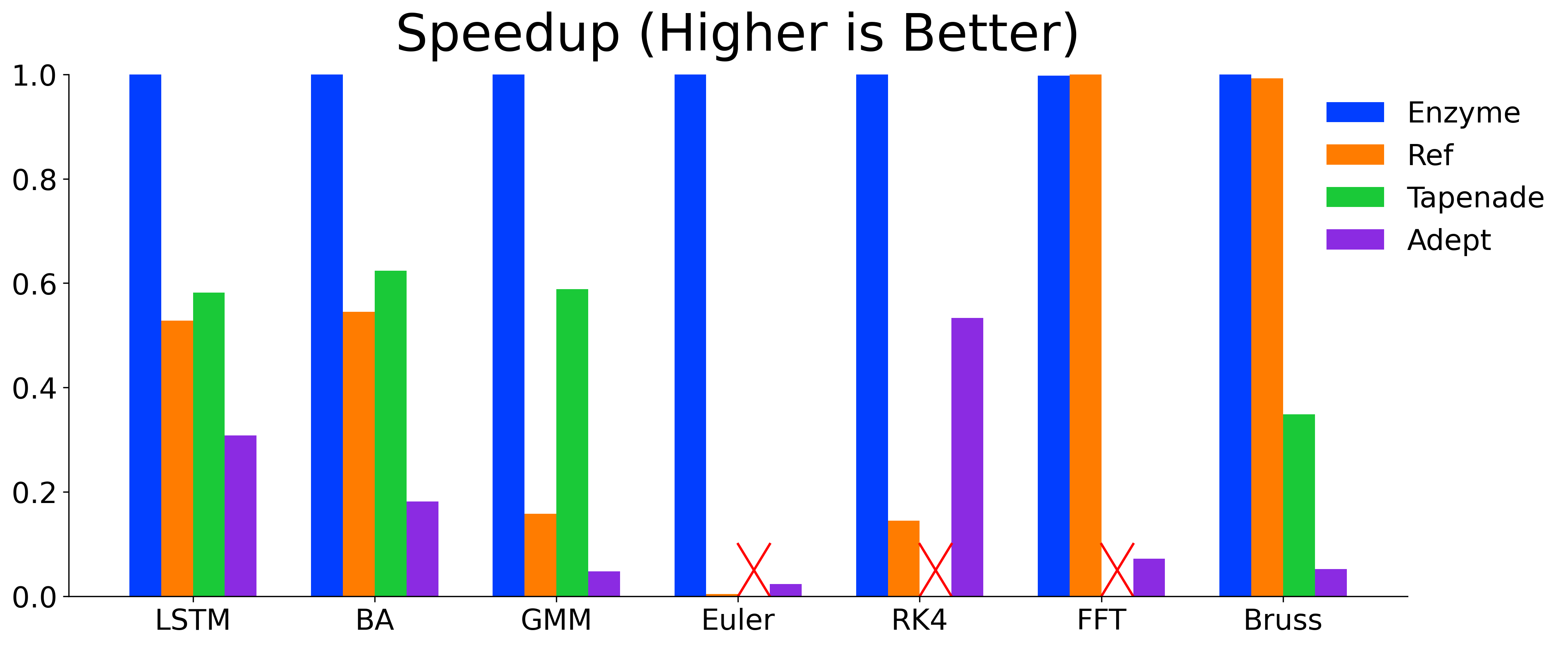
}Enzyme is highly-efficient and its ability to perform AD on optimized code allows Enzyme to meet or exceed the performance of state-of-the-art AD tools.
Detailed information on installing and using Enzyme can be found on our website: https://enzyme.mit.edu.
A short example of how to install Enzyme is below:
cd /path/to/Enzyme/enzyme
mkdir build && cd build
cmake -G Ninja .. -DLLVM_DIR=/path/to/llvm/lib/cmake/llvm -DLLVM_EXTERNAL_LIT=/path/to/lit/lit.py
ninja
Or, install Enzyme using a package manager:
brew install enzyme
spack install enzyme
To get involved or if you have questions, please join our mailing list.
If using this code in an academic setting, please cite the following three papers (first for Enzyme as a whole, second for GPU+optimizations, and third for AD of all other parallel programs (OpenMP, MPI, Julia Tasks, etc.)):
@inproceedings{NEURIPS2020_9332c513,
author = {Moses, William and Churavy, Valentin},
booktitle = {Advances in Neural Information Processing Systems},
editor = {H. Larochelle and M. Ranzato and R. Hadsell and M. F. Balcan and H. Lin},
pages = {12472--12485},
publisher = {Curran Associates, Inc.},
title = {Instead of Rewriting Foreign Code for Machine Learning, Automatically Synthesize Fast Gradients},
url = {https://proceedings.neurips.cc/paper/2020/file/9332c513ef44b682e9347822c2e457ac-Paper.pdf},
volume = {33},
year = {2020}
}
@inproceedings{10.1145/3458817.3476165,
author = {Moses, William S. and Churavy, Valentin and Paehler, Ludger and H\"{u}ckelheim, Jan and Narayanan, Sri Hari Krishna and Schanen, Michel and Doerfert, Johannes},
title = {Reverse-Mode Automatic Differentiation and Optimization of GPU Kernels via Enzyme},
year = {2021},
isbn = {9781450384421},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3458817.3476165},
doi = {10.1145/3458817.3476165},
booktitle = {Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis},
articleno = {61},
numpages = {16},
keywords = {CUDA, LLVM, ROCm, HPC, AD, GPU, automatic differentiation},
location = {St. Louis, Missouri},
series = {SC '21}
}
@inproceedings{10.5555/3571885.3571964,
author = {Moses, William S. and Narayanan, Sri Hari Krishna and Paehler, Ludger and Churavy, Valentin and Schanen, Michel and H\"{u}ckelheim, Jan and Doerfert, Johannes and Hovland, Paul},
title = {Scalable Automatic Differentiation of Multiple Parallel Paradigms through Compiler Augmentation},
year = {2022},
isbn = {9784665454445},
publisher = {IEEE Press},
booktitle = {Proceedings of the International Conference on High Performance Computing, Networking, Storage and Analysis},
articleno = {60},
numpages = {18},
keywords = {automatic differentiation, tasks, OpenMP, compiler, Julia, parallel, Enzyme, C++, RAJA, hybrid parallelization, MPI, distributed, LLVM},
location = {Dallas, Texas},
series = {SC '22}
}
Both Julia bindings and Rust bindings are available for Enzyme.