Official code repository for: FCN-Transformer Feature Fusion for Polyp Segmentation (MIUA 2022 paper)
Authors: Edward Sanderson and Bogdan J. Matuszewski
Links to the paper:
Colonoscopy is widely recognised as the gold standard procedure for the early detection of colorectal cancer (CRC). Segmentation is valuable for two significant clinical applications, namely lesion detection and classification, providing means to improve accuracy and robustness. The manual segmentation of polyps in colonoscopy images is timeconsuming. As a result, the use of deep learning (DL) for automation of polyp segmentation has become important. However, DL-based solutions can be vulnerable to overfitting and the resulting inability to generalise to images captured by different colonoscopes. Recent transformer-based architectures for semantic segmentation both achieve higher performance and generalise better than alternatives, however typically predict a segmentation map of
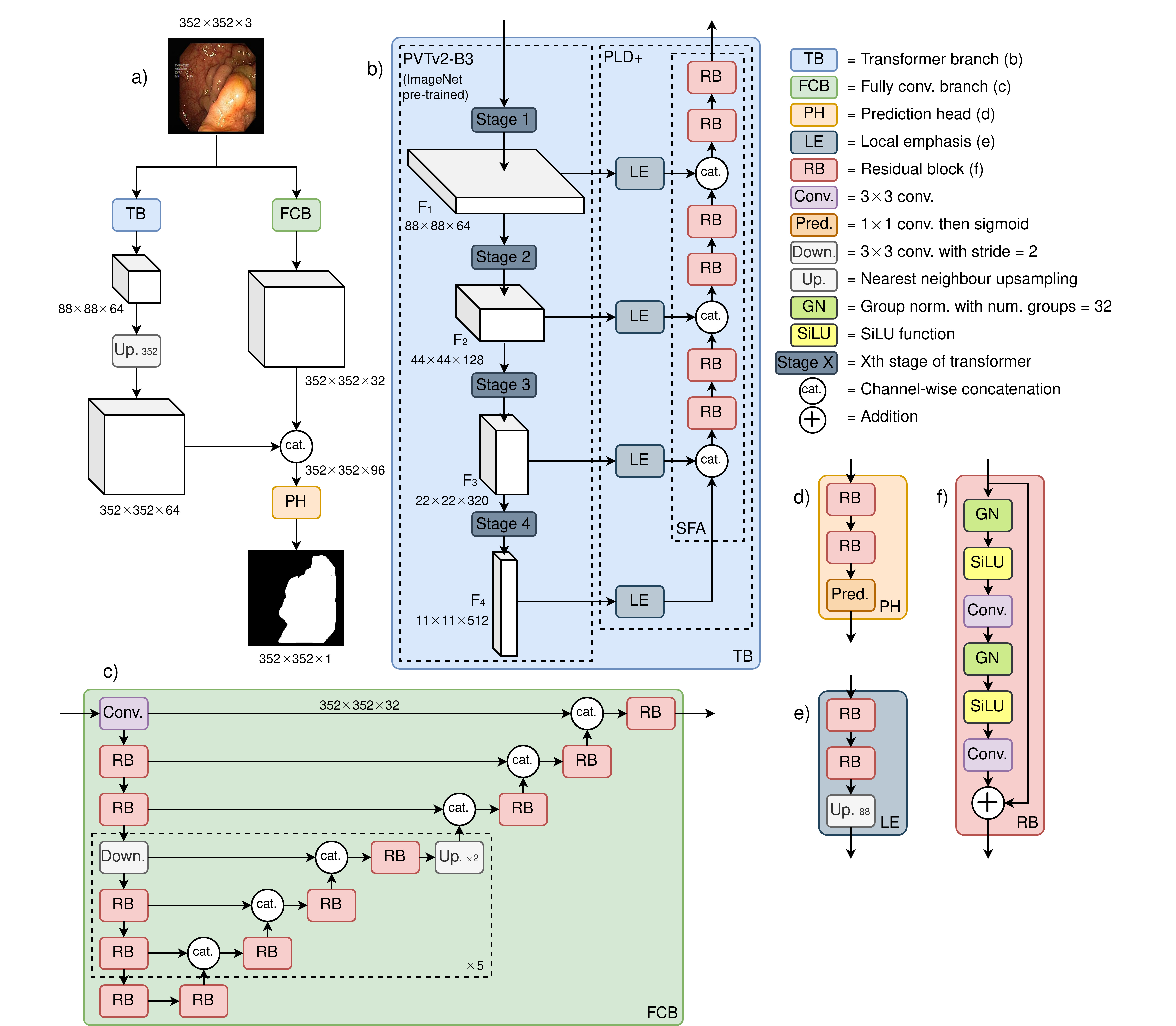
Figure 1: Illustration of the proposed FCBFormer architecture
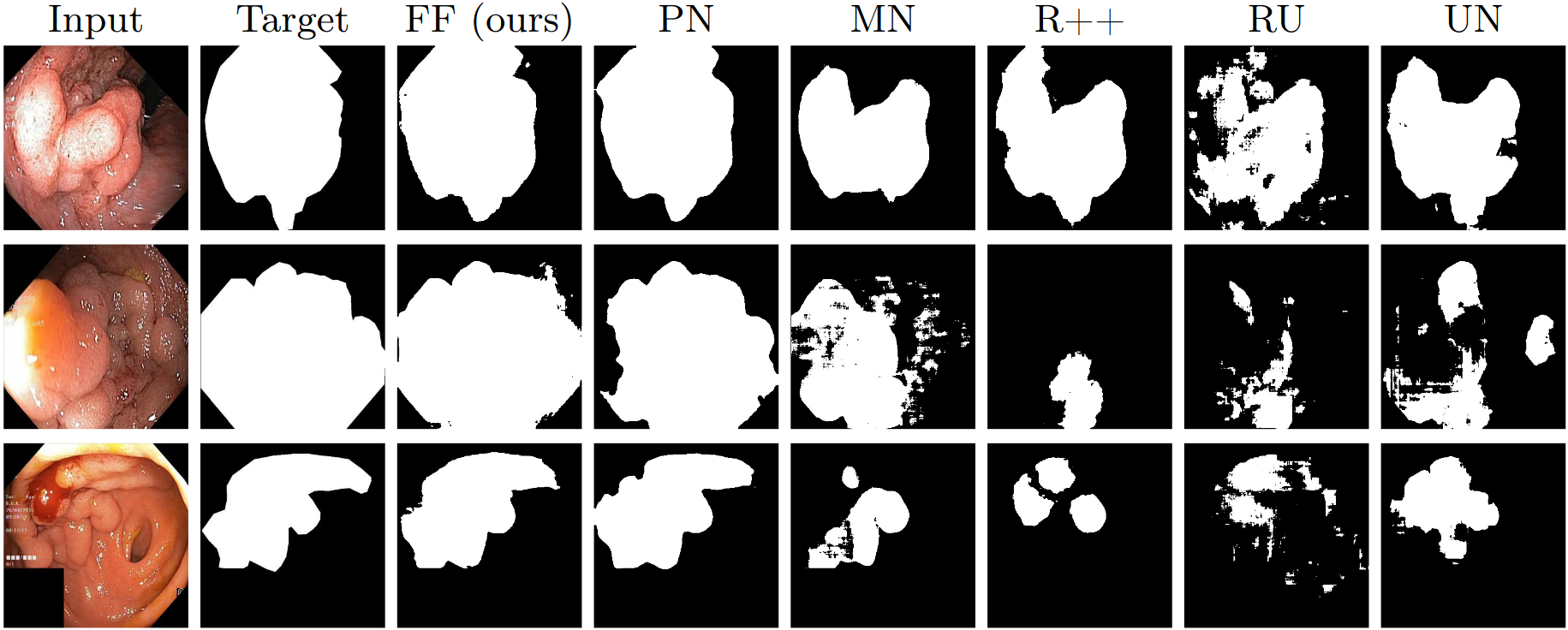
Figure 2: Comparison of predictions of FCBFormer against baselines. FF is FCBFormer, PN is PraNet, MN is MSRF-Net, R++ is ResUNet++, UN is U-Net
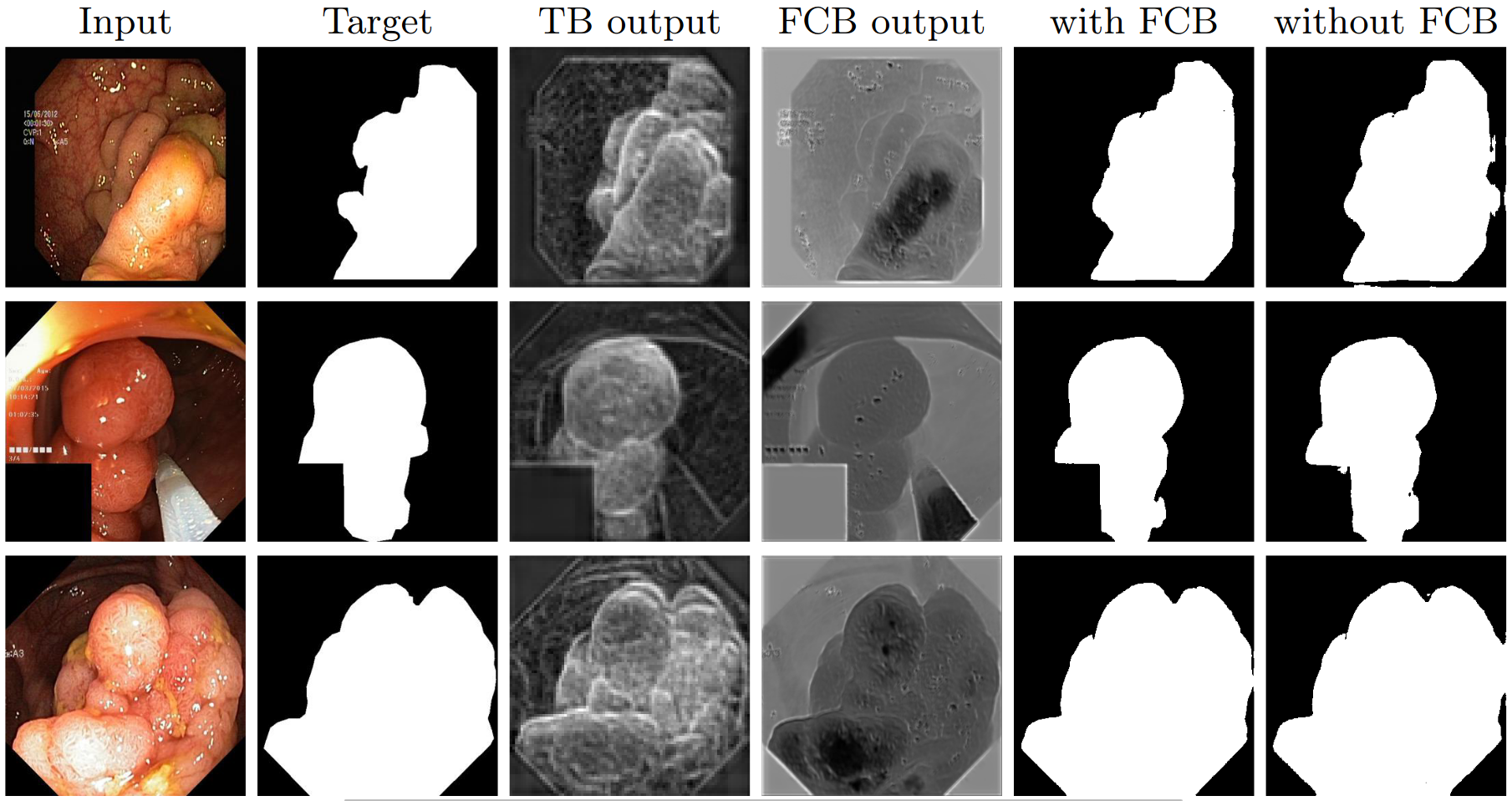
Figure 3: Visualisation of the benefit of the fully convolutional branch (FCB)
- Create and activate virtual environment:
python3 -m venv ~/FCBFormer-env
source ~/FCBFormer-env/bin/activate
- Clone the repository and navigate to new directory:
git clone https://github.com/ESandML/FCBFormer
cd ./FCBFormer
- Install the requirements:
pip install -r requirements.txt
-
Download and extract the Kvasir-SEG and the CVC-ClinicDB datasets.
-
Download the PVTv2-B3 weights to
./
Train FCBFormer on the train split of a dataset:
python train.py --dataset=[train data] --data-root=[path]
-
Replace
[train data]with training dataset name (options:Kvasir;CVC). -
Replace
[path]with path to parent directory of/imagesand/masksdirectories (training on Kvasir-SEG); or parent directory of/Originaland/Ground Truthdirectories (training on CVC-ClinicDB). -
To train on multiple GPUs, include
--multi-gpu=true.
Generate predictions from a trained model for a test split. Note, the test split can be from a different dataset to the train split:
python predict.py --train-dataset=[train data] --test-dataset=[test data] --data-root=[path]
-
Replace
[train data]with training dataset name (options:Kvasir;CVC). -
Replace
[test data]with testing dataset name (options:Kvasir;CVC). -
Replace
[path]with path to parent directory of/imagesand/masksdirectories (testing on Kvasir-SEG); or parent directory of/Originaland/Ground Truthdirectories (testing on CVC-ClinicDB).
Evaluate pre-computed predictions from a trained model for a test split. Note, the test split can be from a different dataset to the train split:
python eval.py --train-dataset=[train data] --test-dataset=[test data] --data-root=[path]
-
Replace
[train data]with training dataset name (options:Kvasir;CVC). -
Replace
[test data]with testing dataset name (options:Kvasir;CVC). -
Replace
[path]with path to parent directory of/imagesand/masksdirectories (testing on Kvasir-SEG); or parent directory of/Originaland/Ground Truthdirectories (testing on CVC-ClinicDB).
This repository is released under the Apache 2.0 license as found in the LICENSE file.
If you use this work, please consider citing us:
@inproceedings{sanderson2022fcn,
title={FCN-Transformer Feature Fusion for Polyp Segmentation},
author={Sanderson, Edward and Matuszewski, Bogdan J},
booktitle={Annual Conference on Medical Image Understanding and Analysis},
pages={892--907},
year={2022},
organization={Springer}
}We allow commerical use of this work, as permitted by the LICENSE. However, where possible, please inform us of this use for the facilitation of our impact case studies.
This work was supported by the Science and Technology Facilities Council [grant number ST/S005404/1].
This work was in part performed using a DiRAC Director’s Discretionary award. The work was carried out on the Cambridge Service for Data Driven Discovery (CSD3), part of which is operated by the University of Cambridge Research Computing on behalf of the STFC DiRAC HPC Facility (www.dirac.ac.uk). The DiRAC component of CSD3 was funded by BEIS capital funding via STFC capital grants ST/P002307/1 and ST/R002452/1 and STFC operations grant ST/R00689X/1. DiRAC is part of the National e-Infrastructure.
This work makes use of data from the Kvasir-SEG dataset, available at https://datasets.simula.no/kvasir-seg/.
This work makes use of data from the CVC-ClinicDB dataset, available at https://polyp.grand-challenge.org/CVCClinicDB/.
This repository includes code (./Models/pvt_v2.py) ported from the PVT/PVTv2 repository.
Links: AIdDeCo Project, CVML Group
Contact: esanderson4@uclan.ac.uk

