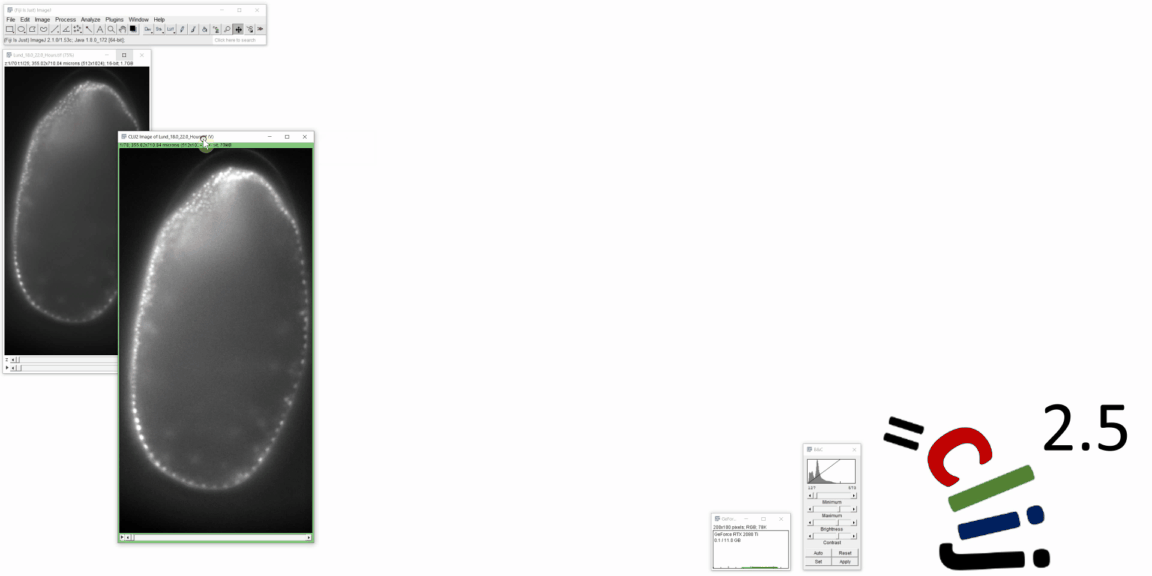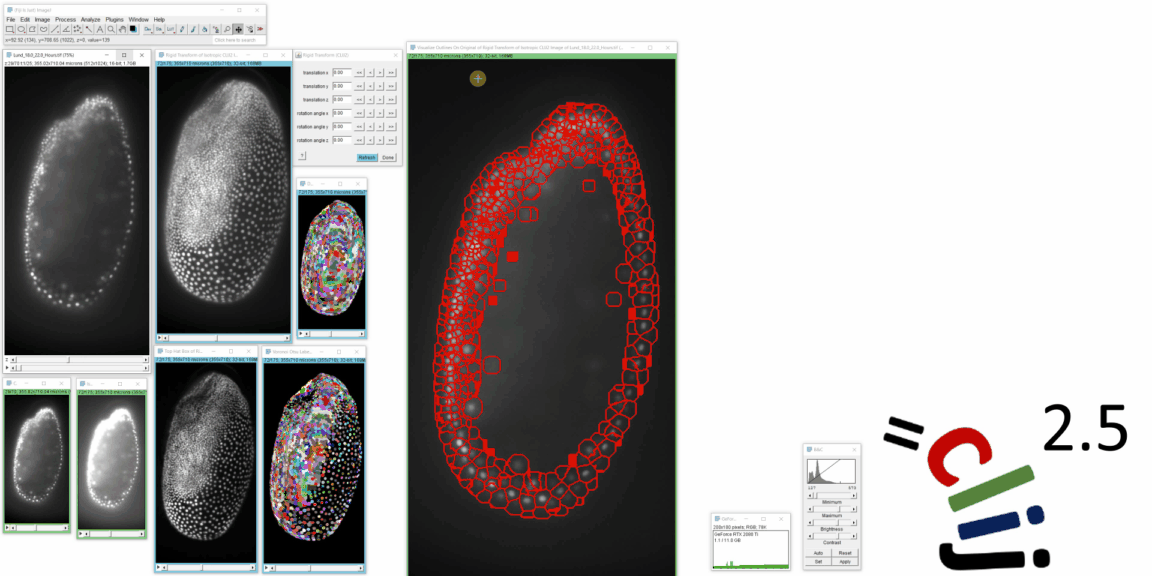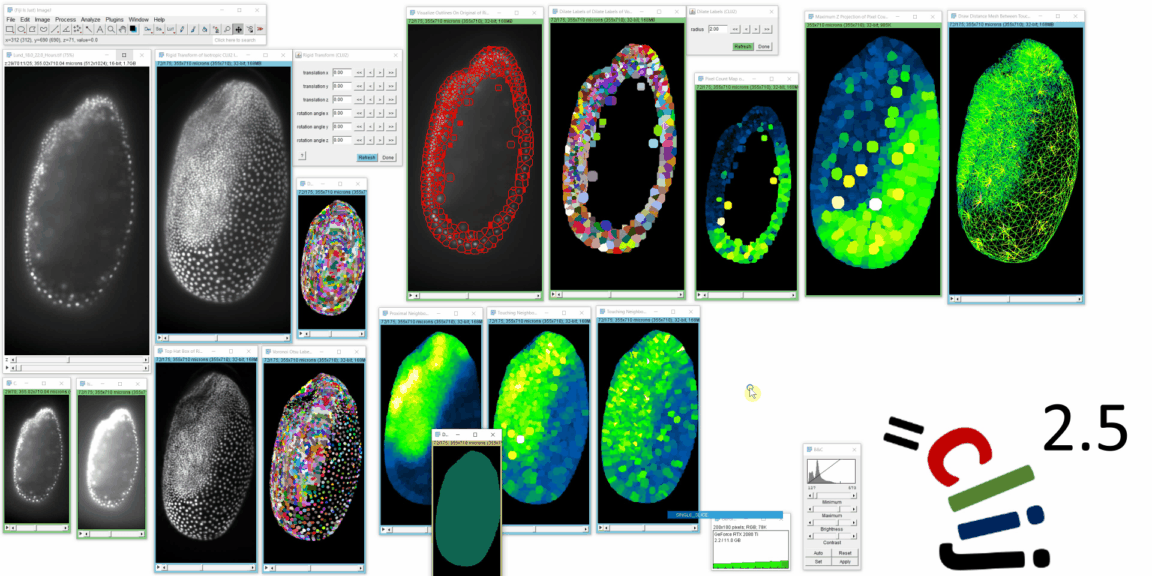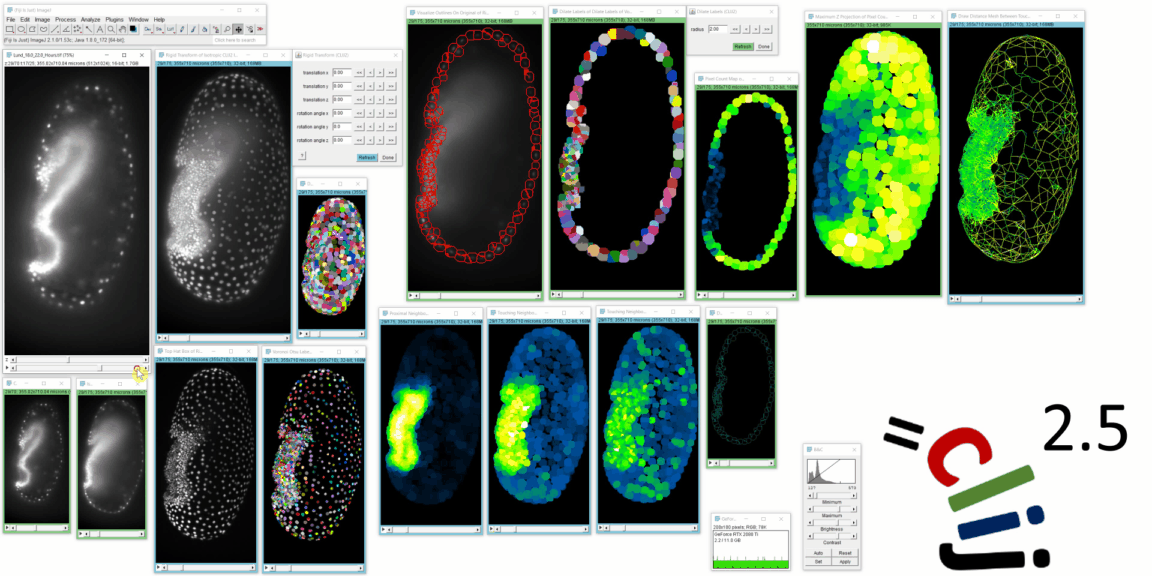
CLIJ2 is a GPU-accelerated image processing library for ImageJ/Fiji, Icy, Matlab and Java. It comes with hundreds of operations for filtering, binarizing, labeling, measuring in images, projections, transformations and mathematical operations for images. While most of these are classical image processing operations, CLIJ2 also allows performing operations on matrices potentially representing neighborhood relationships between cells and pixels.
Under the hood it uses OpenCL but users don't have to learn a new programming language such as OpenCL, they can just use it transparently. Entry-evel coding skills are sufficient! Increased efforts were put on documentation, code examples, user-convenience, interoperability, and extensibility. CLIJ is based on ClearCL, JOCL, Imglib2, ImageJ and SciJava.
CLIJ2 is build on CLIJ. If you use it, please cite it:
-
Robert Haase, Loic Alain Royer, Peter Steinbach, Deborah Schmidt, Alexandr Dibrov, Uwe Schmidt, Martin Weigert, Nicola Maghelli, Pavel Tomancak, Florian Jug, Eugene W Myers. CLIJ: GPU-accelerated image processing for everyone. Nat Methods 17, 5-6 (2020) doi:10.1038/s41592-019-0650-1
-
Daniela Vorkel, Robert Haase. GPU-accelerating ImageJ Macro image processing workflows using CLIJ. arXiv preprint
-
Robert Haase, Akanksha Jain, Stéphane Rigaud, Daniela Vorkel, Pradeep Rajasekhar, Theresa Suckert, Talley J. Lambert, Juan Nunez-Iglesias, Daniel P. Poole, Pavel Tomancak, Eugene W. Myers. Interactive design of GPU-accelerated Image Data Flow Graphs and cross-platform deployment using multi-lingual code generation. bioRxiv preprint
If you search for support, please open a thread on the image.sc forum.
- Introduction to the graphical user interface clij2-assistant
- Installation
- Cheat sheets
- Reference
- Frequently asked questions
- Release notes
- Community guidelines
- Release cycle
- Future perspectives
- Support
- Basics of GPU-accelerated image processing and memory management
- Basic image processing workflows
- Binary images, label images and parametric images
- Introduction for ImageJ Macro users
- Processing multi-channel time-lapse data
- Advanced clij programming
- Image filtering using the assistant
- Gamma correction using the assistant
- Gaussian blur
- Drosophila embryo cell counting
- Crop, Pan & zoom using the assistant
- Cylinder projection using the assistant
- Sphere projection using the assistant
- Crop and paste images
- Maximum projections
- Warp images
- Spot detection
- Nuclei segmentation using the assistant
- Cell segmentation based on membranes using the assistant
- Optimize parameters for binarization using the assistant
- Labeling
- 3D Image Segmentation
- Binary images
- Working with regions of interest (ROIs)
- Adding label outlines and numbers as overlay
- Voronoi diagrams
- Voronoi Otsu labeling
- Classic watershed (MorpholibJ extension)
- Label maps and voronoi diagrams
- Custom clij functions for image segmentation
- Multiply vectors and matrices
- Matrix multiplication
- Spots, pointlists, matrices and tables
- Filtering in graphs
- Filtering between touching neighbors
- Neighbors of neighbors
- Neighborhood definitions
- Tribolium embryo morphometry
- Superpixel segmentation
- Images statistics
- Pixel statistics on labeled images
- Parametric images
- Count neighbors
- Mean of touching neighbors
- Working with tables
- Colocalisation measurements using Jaccard index and Sorensen/Dice coefficient
- Measuring presence of objects between channels
- Measure speedup
- Detailed time tracing
- Comparing and benchmarking workflows
- Comparing image rotation ImageJ vs. CLIJ
- Export workflows as ImageJ Script
- Export human readable protocols and ImageJ Macro Markdown notebooks
- Export as Icy Protocol
- CLIJx-Assistant: My favourite image analysis tool, by Neubias members, by Elnaz Fazeli
- Euro-BioImaging Virtual Pub: Robert Haase, TU Dresden, on "GPU-accelerated image analysis in Fiji and Napari"
- Imaging ONE World: GPU-accelerated 3D image processing for everyone
- I2K2020: Designing GPU-accelerated Image Data Flow Graphs for CLIJ2 and clEsperanto
- GPU-accelerated image processing for the life-sciences, guest lecture at Computer Science Faculty, TU Dresden
- How CLIJ2 can make your bio-image analysis workflows incredibly fast, FocalPlane
- YouTube NEUBIAS Academy @home
- NEUBIAS Symposium 2020, Invited talk, Bordeaux
- NEUBIAS Training School TS14, Teaching session, Bordeaux
- MTZ Image Processing Seminar, Teaching session, TU Dresden
- Quantitative BioImaging Conference 2020, Selected talk, Oxford
- Fast, Faster, CLIJ, News, Center for Systems Biology Dresden
- CLIJ: GPU-accelerated image processing for everyone, Article, Nature Methods
- NEUBIAS Training School TS13, Teaching session, Porto
- ImageJ macro
- Icy javascript
- Icy protocols
- Matlab
- Java
- ImageJ Beanshell
- ImageJ Groovy
- ImageJ Javascript
- ImageJ Jython
- GPU Image Processing using OpenCL, Harald Scheidl, TowardsDataScience
- OpenCL SGEMM tuning for Kepler, Cedric Nugteren, SURFsara
- OpenCL: A Hands-on Introduction, Tim Mattson, Intel
- OpenCL 1.2 Quick Reference, Khronos Group
- OpenCL 1.2 Specifications, Khronos Group
- CLIJ2 - CLIJ2.5 transition guide
- CLIJ versus CLIJ2
- Troubleshooting
- Support
- Imprint
Development of CLIJ is a community effort. We would like to thank everybody who helped developing and testing. In particular thanks goes to Alex Herbert (University of Sussex), Bert Nitzsche (PoL TU Dresden), Bertrand Vernay (IGBMC, Strasbourg) Bram van den Broek (Netherlands Cancer Institute), Brenton Cavanagh (RCSI), Brian Northan (True North Intelligent Algorithms), Bruno C. Vellutini (MPI CBG), Christian Tischer (EMBL Heidelberg), Curtis Rueden (UW-Madison LOCI), Damir Krunic (DKFZ), Daniela Vorkel (MPI CBG), Daniel J. White (GE), Eduardo Conde-Sousa (University of Porto), Elnaz Fazeli (BIU, University of Helsinki), Erick Ratamero (The Jackson Laboratory), Eugene W. Myers (MPI CBG Dresden) Florian Jug (MPI CBG Dresden), Gaby G. Martins (IGC), Gayathri Nadar (MPI CBG Dresden), Guillaume Witz (Bern University), Giovanni Cardone (MPI Biochem), Irene Seijo Barandiaran (MPI CBG Dresden), Jan Brocher (Biovoxxel), Jean-Yves Tinevez (Institute Pasteur), Jim Rowe (University of Cambridge), Johannes Girstmair (MPI CBG), Juergen Gluch (Fraunhofer IKTS), Kisha Sivanathan (Harvard Medical School Boston), Kota Miura, Laurent Thomas (Acquifer), Lior Pytowski (University of Oxford), Marion Louveaux (Institut Pasteur Paris), Matthew Foley (University of Sydney), Matthias Arzt (MPI-CBG), Nico Stuurman (UCSF), Nik Cordes (Los Alamos National Laboratory), Noreen Walker (MPI CBG Dresden), Ofra Golani (Weizmann Institute of Science), Pavel Tomancak (MPI CBG Dresden), Pradeep Rajasekhar (Monash University Melbourne), Patrick Dummer, Peter Haub, Pete Bankhead (University of Edinburgh), Pit Kludig, Pradeep Rajasekhar (Monash University), Rita Fernandes (University of Porto), Romain Guiet (EPFL Lausanne), Ruth Whelan-Jeans, Sebastian Munck (VIB Leuven), Siân Culley (LMCB MRC), Stein Rørvik, Stéphane Dallongeville (Institut Pasteur) Stéphane~Rigaud (Institut Pasteur Paris), Tanner Fadero (UNC-Chapel Hill), Theresa Suckert (OncoRay, TU Dresden), Thomas Irmer (Zeiss), Tobias Pietzsch (MPI-CBG), Wilson Adams (VU Biophotonics)
R.H. was supported by the German Federal Ministry of Research and Education (BMBF) under the code 031L0044 (Sysbio II) and D.S. received support from the German Research Foundation (DFG) under the code JU3110/1-1. P.T. was supported by the European Regional Development Fund in the IT4Innovations national supercomputing center-path to exascale project, project number CZ.02.1.01/0.0/0.0/16_013/0001791 within the Operational Programme Research, Development and Education. We also acknowledge the support by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germanys Excellence Strategy EXC2068 - Cluster of Excellence Physics of Life of TU Dresden. This project has been made possible in part by grant number 2021-237734 (GPU-accelerating Fiji and friends using distributed CLIJ, NEUBIAS-style, EOSS4) from the Chan Zuckerberg Initiative DAF, an advised fund of the Silicon Valley Community Foundation.