Climo is an open-source package that lets you quickly build an online interface to your R models. Deployed models can be freely shared with others so that anyone can explore your model’s predictions. You can also retrieve and validate other interesting models locally on your data.
The features of climo are specifically geared towards models with a clinical medicine focus. We believe that collaboration within the medical community can be improved by focusing on sharing models rather than datasets.
This short overview of the climo package will go through installation and creating a model that can be visualized at clinicalmodels.io.
To start, you can install the development version of climo with the following statement:
devtools::install_github("climo-ai/climo-r")Additionally, to use most of the climo R functions you will need to retrieve your API key from the clinicalmodels.io platform and set it in your R environment. After signing in at clinicalmodels.io, navigate to Home > Profile and copy the API key. Then, run the following command in R to make your key available for the climo package:
Sys.setenv('CLIMO_API_KEY' = '__your key__')If you want the key to persist between R sessions, you can set the key
in your R environment by running usethis::edit_r_environ() and adding
the line CLIMO_API_KEY="__your key__" to the .Renviron file which
gets opened. That way you wont have to set the API key every time you
start R.
You can check that your API key is correctly set by running
Sys.getenv('CLIMO_API_KEY')
Now, let’s create a model that can be uploaded to clinicalmodels.io using the example dataset included in the climo package:
library(climo)
library(nlme)
model <- nlme::lme(CDRSB ~ CDRSB_bl + AGE + GENDER + TIME*CDRSB_bl,
random = ~ TIME | ID,
control = nlme::lmeControl(
maxIter = 1e10,
msMaxIter = 1000,
opt = "optim"
),
data = climo::example,
na.action = stats::na.omit)This lme model can then be used to create a climo model. All that is
needed is to call the create_model function and pass in the minimum
required arguments of the model object, a name for the model, and the
clinical area to which the model belongs (here, Alzheimer’s Disease):
climo_model <- climo::create_model(model, name="example-model', area='Alzheimers Disease')And just like that, we now have a model upload to clinicalmodels.io. To
visualize the model interactively, we can go to clinicalmodels.io and
visit our model page, where we are able to add model inputs from the
settings page. Alternatively, we can add inputs directly from the climo
package as shown in the Create your own model exmaple.
The model inputs make up the interactive user interface to your model on the clinicalmodels.io platform. They’re necessary if you want users to be able to interact with your model at clinicalmodels.io.
There are two types of model inputs: continuous inputs and categorical inputs.
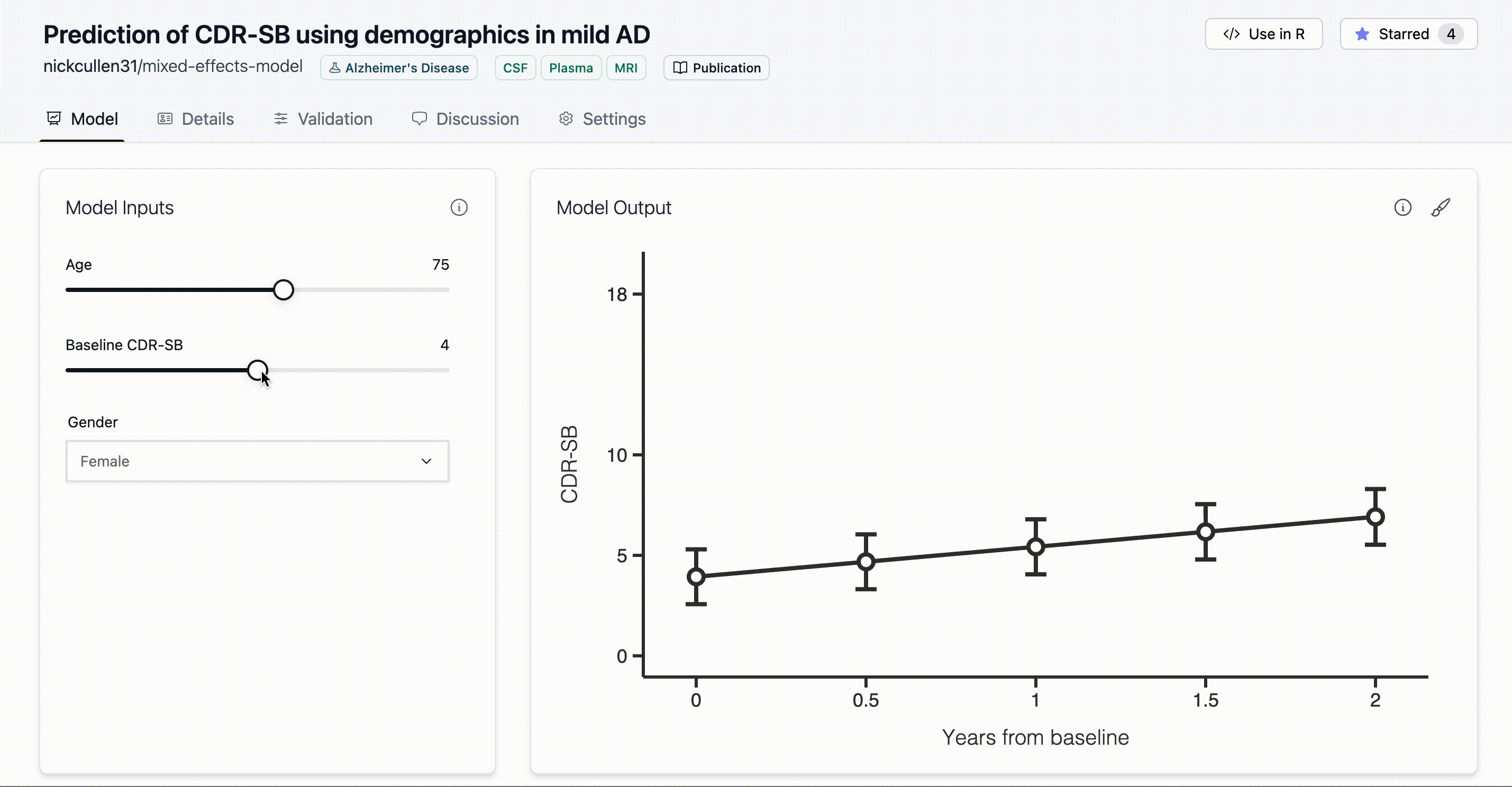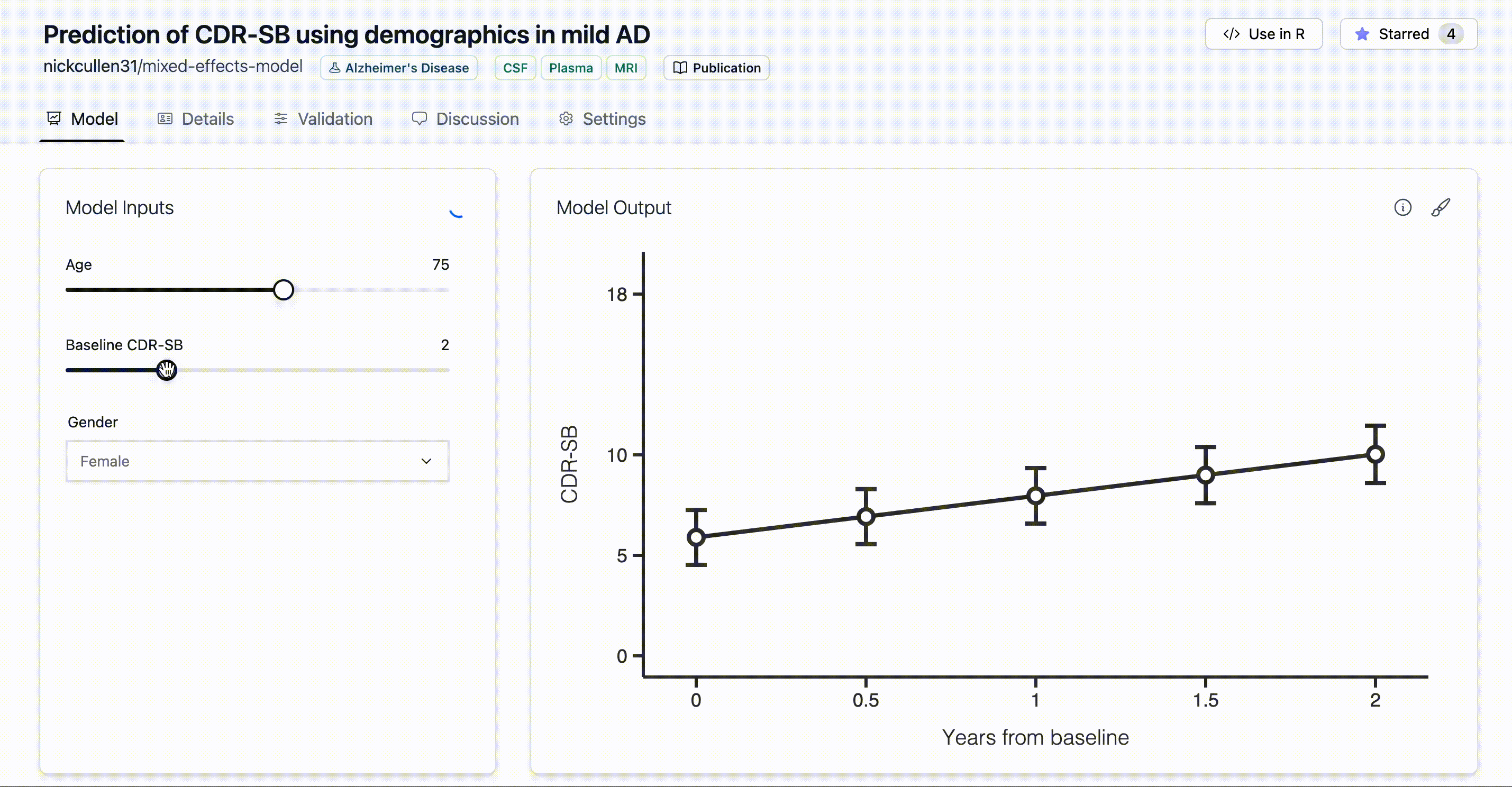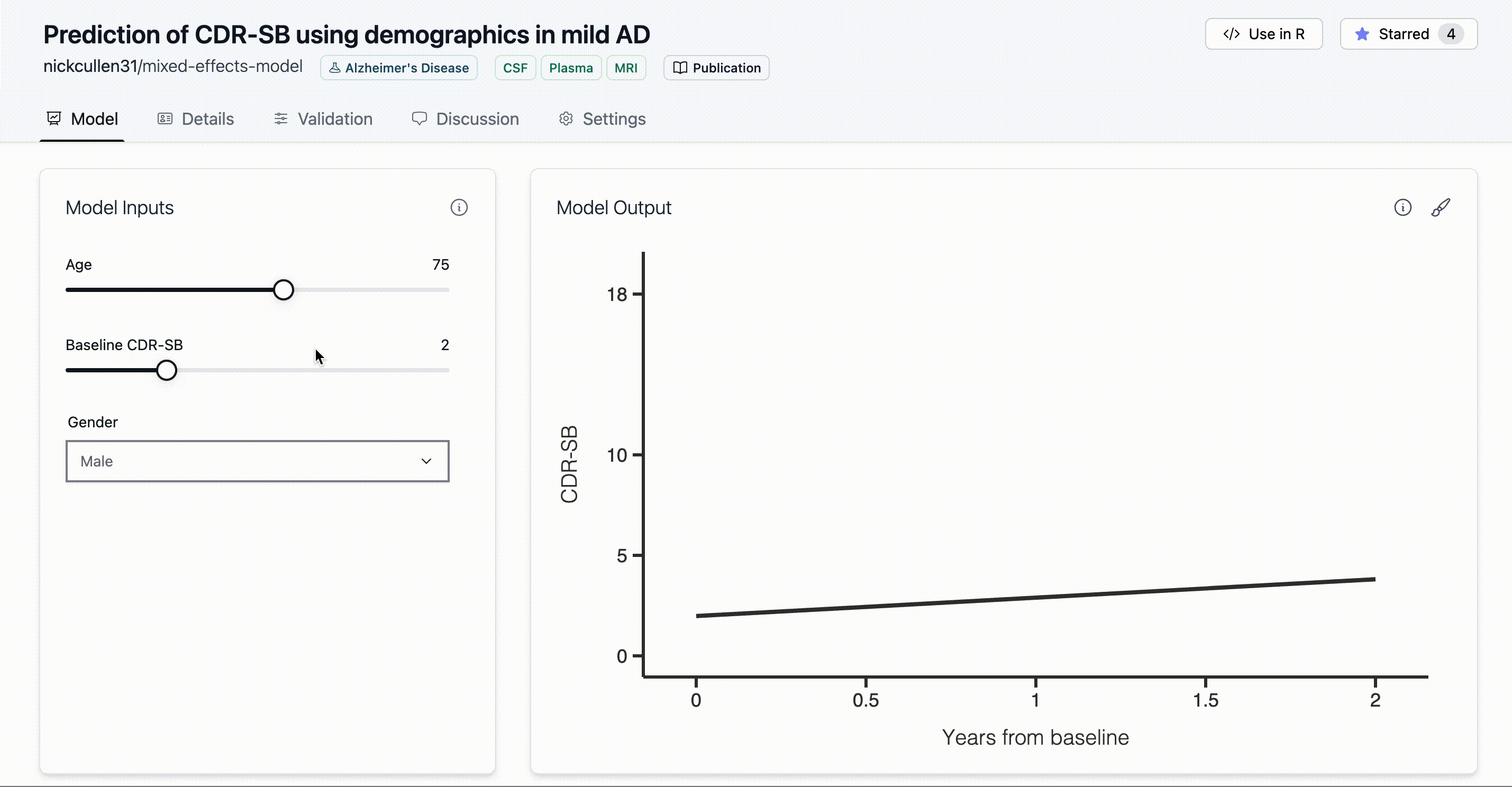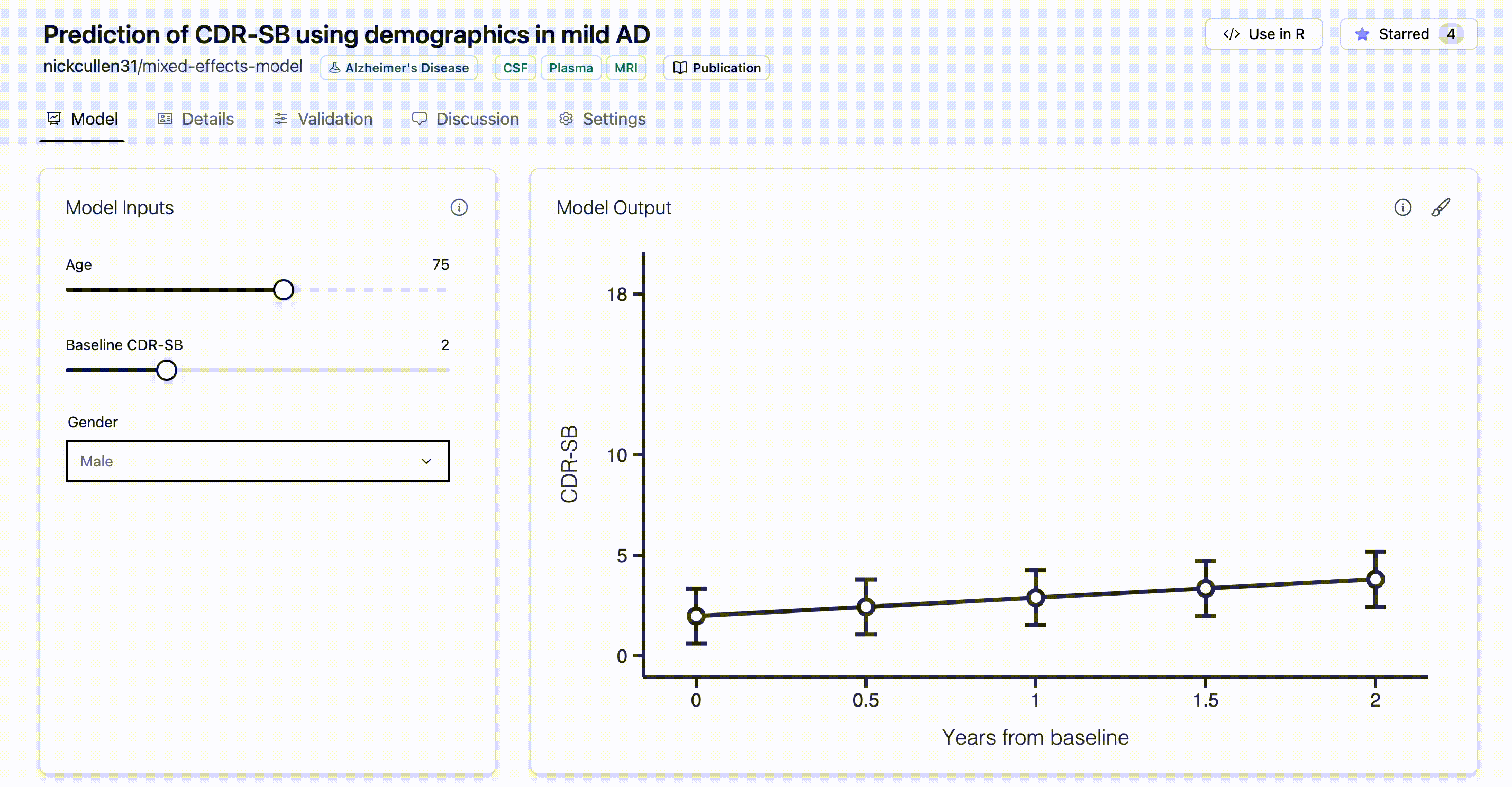
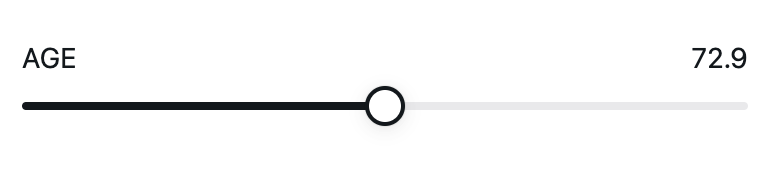
Continuous inputs are represented by sliders for numeric variables, as shown here:
There are a couple of continuous input variables in our model (AGE and
CDRSB_bl), so we can create them as follows:
age_input <- climo::create_input('AGE', label='Age', type='continuous', min=50, max=90, step=1, initial=70)
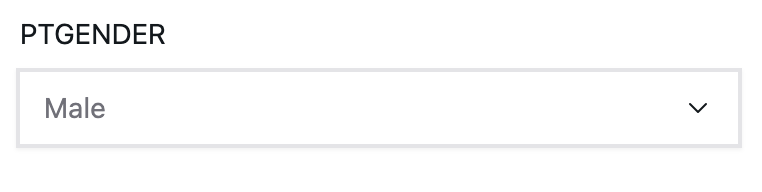
cdrsb_input <- climo::create_input('CDRSB_bl', label='Baseline CDR-SB', type='continuous', min=0, max=10, step=0.5, initial=2.5)Categorical inputs, on the other hand, are represented by dropdowns for discrete variables.
There is also one categorical input variable in the model which we can create in a similar way:
gender_input <- climo::create_input('PTGENDER', label='Gender', type='categorical', options=c('Male','Female'), initial='Female')You may have noticed that the time variable from the model was not
assigned an input. That’s by design – for longitudinal models, you
usually want to fix the time values in order to display a disease
progression curve over time. Therefore, you should add the time variable
as a continuous input, but give it multiple values and specify that it
represents the time variable.
time_input <- climo::create_input('time', label='Years from baseline', type='continuous', options=c(0, 0.5, 1, 1.5, 2), is_time=TRUE)Finally, we have the inputs for all of the five variables in the model and we can actually add them to the model.
climo_model %>% add_inputs(
age_input,
cdrsb_input,
gender_input,
time_input
)By adding the inputs to the model, they will actually show up at clinicalmodels.io and users can interact with the model. You can see the example at clinicalmodels.io/climo/example-r-lme
And just to note, if you ever lose track of your model then you can always retrieve it back from the platform:
climo_model <- climo::retrieve_model('climo/example-r-lme')The platform is smart enough to know which models are yours and which aren’t based on your API key – so no one else besides you can edit your models in any way.
Now that the model has inputs, it is time to specify the display. The display parameters control how the acutal figure that gets plotted looks like. At this time, there is only one parameter available: the output label which will serve as the y-axis label. Adding the display is simple:
climo_model %>% add_display(output_label = 'CDR-SB')Now, when the figure is plotted it will show the correct output label rather than the default value of “Output”.
The model visualization is now complete, but there is still more to do. It’s important to provide details about an uploaded model so that other users can get an idea of how the model was fit, what its limitations are, and what the appropriate context for the model is. This information can be provided in the model details, which is by default composed of four sections:
- Participants
- Outcome
- Predictors
- Methods
The participants section should fully describe the number and characteristics of the individuals whose data was used when fitting the model, along with any other important information about the study cohort. The outcome section should describe how the outcome variable was collected and its relevance for the disease. Similarly, the predictors section should describe how all of the predictors (or inputs or covariates - however you would like to call them) were collected. Finally, the methods section should give a full description of the model itself – how it was fit and evaluated, and what software packages were used.
Following the same pattern as before, the model details can be added like this:
climo_model %>% add_details(
participants = "This is the participants section",
outcome = "This is the outcome section",
predictors = "This is the predictors section",
methods = "This is the methods section",
)Now, we have a model on the clinicalmodels.io platform which is
available for users to interactive visualize and which also includes key
details about the model. The final model can be visited at
www.clinicalmodels.io/{your_username}/example-model.
Besides visualizing and describing models on the clinicalmodels.io platform, it is also possible to evaluate models. A model should always be internally validated using the data on which the model was trained, but evaluating models on external datasets is also worthwhile.
If you have a cohort dataset, feel free to explore other models on clinicalmodels.io which you are able to externally validate. This improves the credibility and validity of clinical predictions models.
Validating models from the climo package is quite straight-forward. First, start by retrieving the model which you want to validate. We can use the model we created previously to show how an internal validation can be performed.
library(climo)
model <- retrieve_model('nickcullen31/example-prognostic-model')To understand what variables the model expects, we can print out the inputs to the model:
print(model$inputs)However, since we are validating the model on the same data which we used to fit the model, we do not need to worry about changing any variable names. Now, we call the function to evaluate the model on the dataset:
results <- evaluate_model(model, newdata = climo::example_data)Notice that we used the climo::example_data dataset which is included
in the climo package, but you would pass in whatever dataframe you used
to fit the original model.
Additionally, you may notice that we didn’t specify any methods to evaluate the model. In fact, climo knows from the model type how it should be evaluating. In the case of our mixed-effects model with a continuous outcome, the R^2 value will be calculated.
Finally, we can submit our validation to the clinicalmodels.io platform so that the community can see that we have appropriately validated our model.
model %>% add_validation(results, cohort='Training Data', internal=TRUE)The R^2 value will show up on our model’s Validation page, as will two figures: a scatter plot of the observed outcome values versus the predicted outcome values, and a box plot showing the distribution of error values (predicted - observd values).
As other people validate your model with external data, those results will also show up in a nice comparison table and figure on your model’s validation page.
If you have any questions, feel free to submit an issue here on GitHub or reach out to us at info@clinicalmodels.io.