The Multiple Myeloma Research Foundation (MMRF) CoMMpass registry contains longitudinal data for over 1000 newly diagnosed multiple myeloma patients. Researchers track these patients from initial diagnosis through their course of treatment over a minimum of 5 years, capturing patient lab values, treatments, and complications. In addition to these data, there is a rich store of biological and demographic data of each patient at baseline, including RNA-seq, cytogenetics, and survey (e.g. Quality of Life) data.
ML-MMRF is a repository built to process the MMRF CoMMpass Dataset and allows researchers to use these data for machine learning. We first parse the raw MMRF files into tensors (stored in numpy matrices), then clean and normalize the tensors, and finally provide a notebook for validation of the procedure.
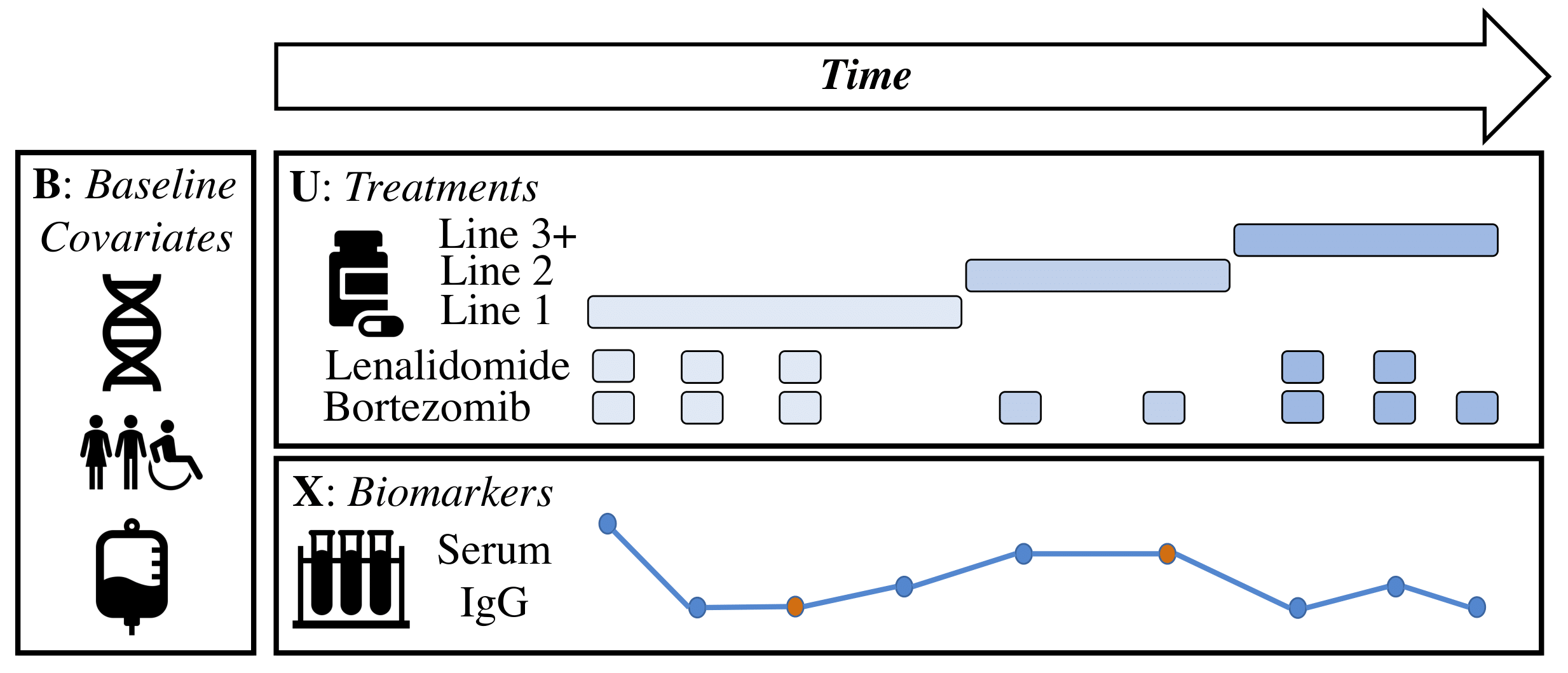 Figure 1 - Example of Patient Data Post-Processed w/ ML-MMRF: Illustration of data from a multiple myeloma patient. Baseline (static) data typically consists of genomics, demographics, and initial labs. Longitudinal data typically includes laboratory values (e.g. serum IgG) and treatments. Baseline data is usually complete, but longitudinal measurements are frequently missing at various time points. The data tells a rich story of a patient's disease trajectory and the resulting treatment decisions. For example, a deviation of a lab value from a healthy range (e.g. spike in serum IgG) might prompt a move to the next line of therapy. Missing data (e.g. points in red) in this case are forward filled.
Figure 1 - Example of Patient Data Post-Processed w/ ML-MMRF: Illustration of data from a multiple myeloma patient. Baseline (static) data typically consists of genomics, demographics, and initial labs. Longitudinal data typically includes laboratory values (e.g. serum IgG) and treatments. Baseline data is usually complete, but longitudinal measurements are frequently missing at various time points. The data tells a rich story of a patient's disease trajectory and the resulting treatment decisions. For example, a deviation of a lab value from a healthy range (e.g. spike in serum IgG) might prompt a move to the next line of therapy. Missing data (e.g. points in red) in this case are forward filled.
Access to the MMRF CoMMpass data is through the MMRF Researcher Gateway. To request access, you should e-mail Jen Rogers rogersj@themmrf.org. Once you have access, please download the FlatFiles and the associated dictionaries as well as the file, MMRF_CoMMpass_IA15_Seq_QC_Summary.xlsx and MMRF_CoMMpass_IA15a_E74GTF_Cufflinks_Gene_FPKM.txt. The latter files will be necessary for processing of the genetic data.
- Enter the folder ml_mmrf.
- Run the code in
requirements.shto setup the relevant packages you will need in order to setup the data. - Sign up on the MMRF Gateway linked above to receive access to the data and then download FlatFiles.
- Finally, go to the
corefolder and run:python build_mmrf_dataset.py --fdir [PATH TO FLATFILES] --outcomes_type [OUTCOMES_TYPE (mortality or trt_resp)] --ia_version IA15 --recreate_splits Falseto create the data tensors from the raw MMRF flatfiles. Note that there are additional arguments you can specify, although this is not strictly necessary. Please see the script for details.
After data processing, we recommend going through the provided jupyter notebook, "3_SanityCheckData.ipynb" to verify that the tensors have been created properly. Below, we detail the specific aspects of the outer level build script.
The Parser class in core/parser.py is responsible for taking the raw data stored in pandas dataframes and converting it into sequential data tensors of size N x maxT x D, where N is the number of examples, maxT is the max number of time steps, and D is the feature dimension. The user specifies the granularity of time at which we parse, i.e. the number of days between subsequent time steps, and also specifies maxT, the max time for which we parse. Finally, we also return a binary mask tensor, where we store a value of 1 if it is observed and 0 if it is missing. As an example, if we specify granularity to be 60 and maxT to be 33, which are the default settings, then the treatment and labs tensors will be of size N x 33 x D. Furthermore, suppose t = 1,...,maxT; the time between t and t+1 is determined by granularity, which in this case is 60 days (2 months).
There are three data types that we parse from the raw MMRF files: treatments, patient labs, and baseline data (basic demographics and cytogenetics). We detail the specific features available in the raw files in the Data Description section below. For now, we will give a brief overview of how we actually do the parsing for each data type.
The subset of lab values that we select from the raw files at each visit include a patient's blood chemistry values (i.e. albumin, BUN, calcium, creatinine, glucose, and total protein), complete blood counts (i.e. absolute neutrophils, hemoglobin, WBC, and plateletes), and finally, their serum immunoglobuins (i.e. IgG, IgA, IgM, lambda, kappa, and M-protein). We then clip the values of these labs to 5 times the median value, and then build the sequential tensor by processing the values based on the user-specified granularity and maxT. Each visit and lab collection panel is labeled with the day in which it was taken, allowing us to bucket the lab value into a particular time step. The lab values at a patient's baseline visit are mean imputed across the cohort at baseline.
We obtain the treatments given to a patient across time along with the line of therapy that each treatment is associated with. Additionally, we restrict the treatments to those that appear in the top 10 treatment combinations with respect to raw counts over the entire time course. The final treatment representation that we use is a 9-dimensional binary vector where five dimensions refer to whether or not one of Dexamethasone, Lenalidomide, Bortezomib, Carfilzomib, and Cyclophosphamide are given. The sixth thru ninth dimensions of the binary vector are a one-hot representation of the line of therapy, which we categorize into one of three buckets: Line 1, Line 2, or >= Line 3. When parsing the treatments, we leverage the start and end days which are given for each of the regimens to construct the tensor, whose final size is N x maxT x 9.
We extract several features that are gathered for each patient at baseline (i.e. before they are started on their treatment course). These include demographic features, such as age and gender, multiple myeloma subtype, namely IgG type, IgA type, which we compute based on serum lab values at baseline, and the PCA output of patient RNA-seq data. We do mean imputation on all missing baseline data. However, for the genetic PCA data, we do knn (k=5) imputation.
Tasks of potential clinical interest in multiple myeloma often include predicting an outcome for a patient conditioned on their treatments, disease subtype, genetic information, and lab values. These clinical outcomes include time to death, 1-year or 2-year mortality, and treatment response to a particular line of therapy. Of note, treatment response in multiple myeloma is categorized into six buckets from most responsive to least responsive: 'stringent complete response', 'complete response', 'very good partial response', 'partial response', 'stable disease', and 'progressive disease'. The Parser class collects the second line treatment response for each patient as a binary variable between 'progressive disease' and non-'progressive disease'. It also collects the time to death for each patient. We restrict the number of patients in our cohort based on who has an observed clinical outcome of interest (specified by user).
The Cleaner class in core/cleaner.py is responsible for taking the tensors outputted by the Parser above and normalizing the values. Normalization procedure, which we describe below, is different for each data type. For example, we z-score normalize each of the baseline features. Namely, for a baseline feature, b, we have normb = (b - mean(b)) / std(b). For the lab values, we utilize a slightly different normalization procedure. We normalize such that the value being >0 refers to an unhealthy lab. Specifically, we subtract the healthy maximum value and then multiply the lab by a scaling factor to map it to a reasonable range (<10).
The Splitter class in core/splitter.py is responsible for generating the train and test sets for the MMRF cohort. It also generates k folds (k specified by the user but generally we have used k=5) of the training data for k-fold cross validation. The split is balanced on the clinical outcome specified by the user above.
The notebook, "3_SanityCheckData.ipynb", allows a user to see if the labs, treatments, and baseline data were processed correctly.
There are three main non-genetic data files in CSV format that we use, which are included in the FlatFiles released by the MMRF:
MMRF_CoMMpass_IA15_PER_PATIENT.csv: Contains the baseline demographic (age, gender, race, and ethnicity) and cytogenetics (e.g. FISH results) for each patient. It also includes ISS stage.MMRF_CoMMpass_IA15_PER_PATIENT_VISIT.csv: Contains the sequential lab values and other longitudinal information about each patient. Specifically, we are given the visit date, any labs collected during that visit, as well as symptoms and signs gathered from physical exam, patient history, and/or labs (e.g. fatigue, bone pain, hypercalcemia, etc.). We are also given quality of life survey data.MMRF_CoMMpass_IA15_STAND_ALONE_TRTRESP.csv: Contains the longitudinal treatment information for each patient, including the therapy name and class, start and end days for each therapy, the line associated with the therapy, and the best response for each line.MMRF_CoMMpass_{ia_version.upper()}a_E74GTF_Cufflinks_Gene_FPKM.txt: Contains the RNA-seq data for each patient.GRCh37.p13_E92_mart_export.txt: Genome reference assembly file that doesn't come with the MMRF Study but that we provide. Can also be downloaded from here.
Each flat file is associated with a data dictionary upon download. Each dictionary has three relevant columns: name, which has the feature names, label, which contains a short description of the feature, and vartype, which details the type of feature (e.g. CHAR, NUM).