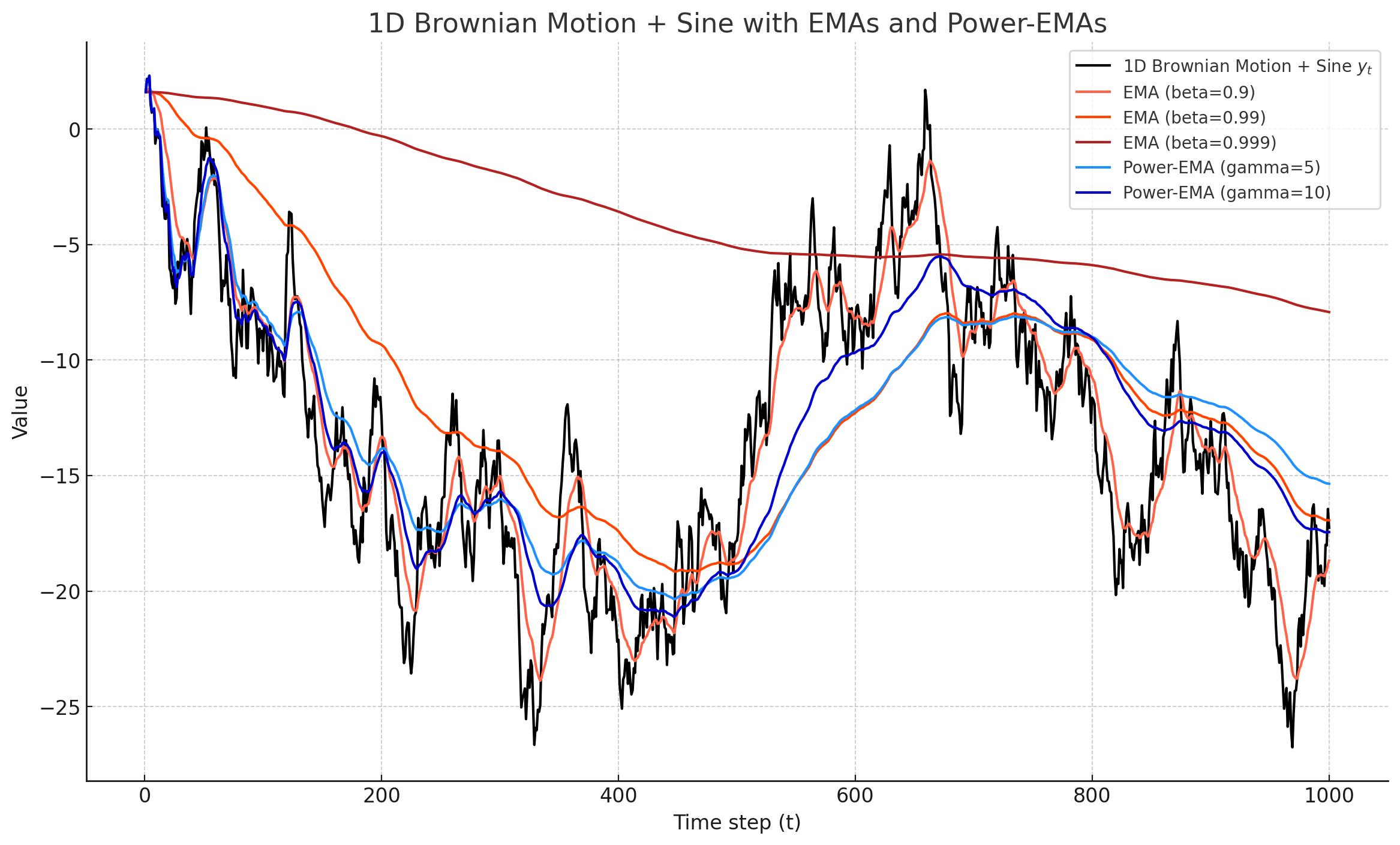
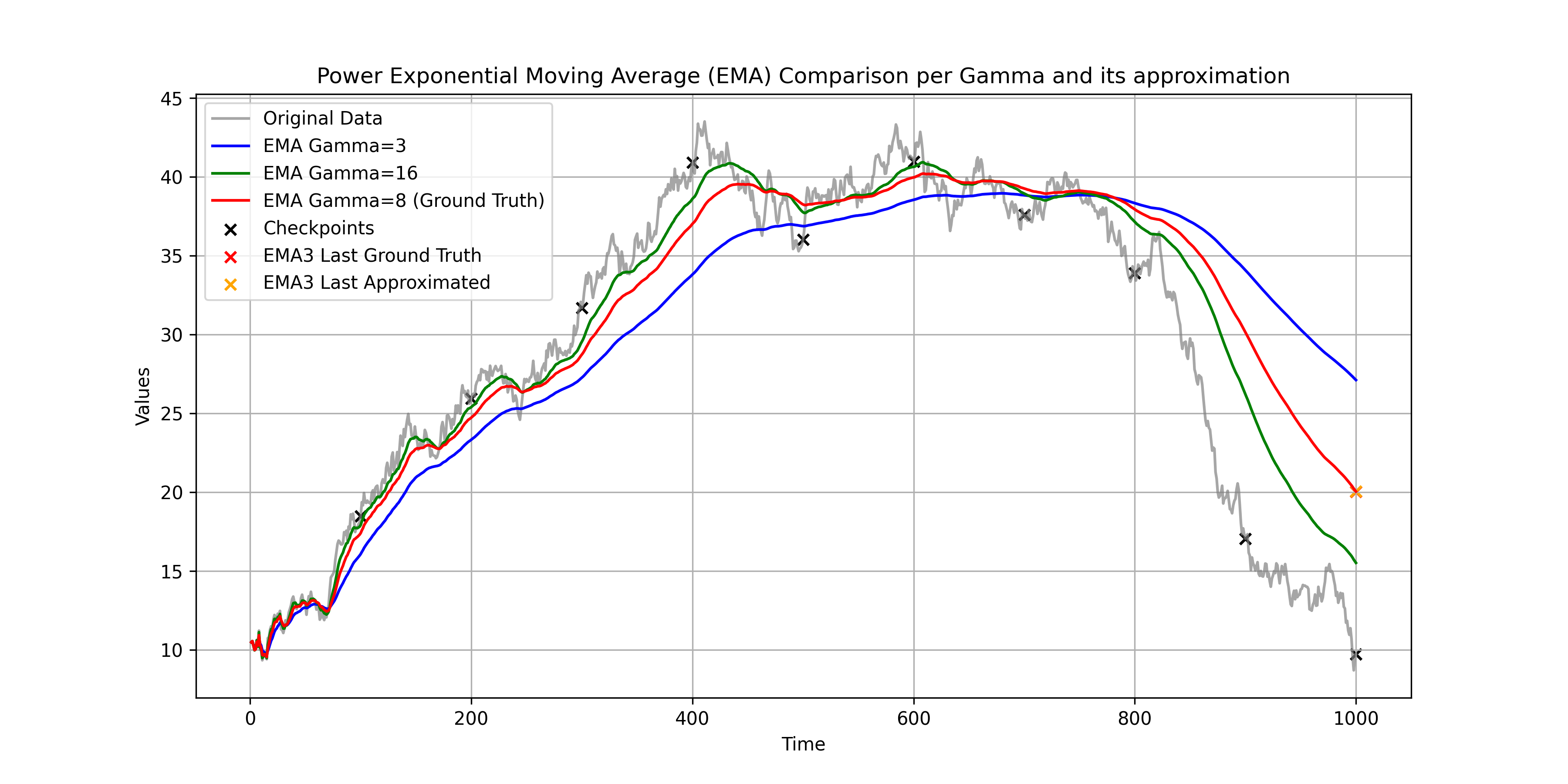
This tutorial-repo implements the Karras's Power function EMA, quite incredible trick introduced in the paper Analyzing and Improving the Training Dynamics of Diffusion Models by Tero Karras, Miika Aittala, Jaakko Lehtinen, Janne Hellsten, Timo Aila, Samuli Laine.
I recommend you to read the paper for full detail, but here is the big picture.
Recall that EMA'ing checkpoint is about keeping track of smooth-version of model parameters,
, where
You want to use EMA, but...
- You don't want the ema to be too slow, because it will make random initialization's contribution to the final model too big.
- You definitely want the decaying factor to be self-similar, because you should be able to increase-time of the training.
- You want to set decaying factor post-hoc, because you don't want to retrain the model from scratch with different decaying factor.
Karras's Power function EMA is the answer to all of these problems. He first uses power-function version of EMA where instead of keeping beta constant, he uses
So there is two main part of the algorithm.
- Saving two copies of the EMA-model, each with different width.
- Recovering arbitrary-width EMA
Think of width as decaying factor. Larger width means it will be smoother.
This is the easy part. You just need to save two copies of the EMA, each with different width (different
gamma_1 = 5
gamma_2 = 10
model = Model()
model_ema_1 = copy.deepcopy(model).cpu()
model_ema_2 = copy.deepcopy(model).cpu()
for i, batch in enumerate(data_loader):
beta_1 = (1 - 1/(i+1)) ** (1 + gamma_1)
beta_2 = (1 - 1/(i+1)) ** (1 + gamma_2)
# train model
loss.backward()
optimizer.step()
for p, p_ema_1, p_ema_2 in zip(model.parameters(), model_ema_1.parameters(), model_ema_2.parameters()):
p_ema_1.data = p_ema_1.data * beta_1 + p.data * (1 - beta_1)
p_ema_2.data = p_ema_2.data * beta_2 + p.data * (1 - beta_2)
if i % save_freq == 0:
torch.save(model_ema_1.state_dict(), f'./model_ema_1_{i}.pth')
torch.save(model_ema_2.state_dict(), f'./model_ema_2_{i}.pth')Now what if you want to recover EMA with
EMA, by definition, can be considered as integral of trajectory of the model parameters. So if you have some weighting function
For a fixed training runs,
for
where
Our goal is then to
-
find a approximate
$\hat{w}_3(t)$ that will give us the EMA that corresponds with$\gamma_3$ . -
find the correpsonding
$\theta_{3,T}$
See where this is going? Our goal is to approximate
where
Aha! Now we can find
We have
How would you solve this?
Define inner product as
Then we can rewrite the problem as
if we define
Ha, so substituting
where
So the solution is simply
where
Note : Well if you ever studied functional analysis, you realize hey, there exists unique solution to this problem, via Hilbert's Projection Theorem. The above is simply finding the projection of
$g$ onto the subspace spanned by$f_i$ , in$L^2$ space.
So thing you learned:
- The level of approximation is determined by the number of checkpoints you saved. More checkpoints, better approximation.
- This doesn't have to be power-function EMA. You can use any weighting function
$w(t)$ , as long as you can compute the integral of the trajectory of the model parameters.
Ok, but reminder this is just for a power-function EMA. You can use this for any weighting function
In the above code, you saved two copies of EMA, each with different
t_checkpoint = t[checkpoint_index]
ts = np.concatenate((t_checkpoint, t_checkpoint))
gammas = np.concatenate(
(
np.ones_like(checkpoint_index) * gamma_1,
np.ones_like(checkpoint_index) * gamma_2,
)
)
x = solve_weights(ts, gammas, last_index, gamma_3)
emapoints = np.concatenate((y_t_ema1[checkpoint_index], y_t_ema2[checkpoint_index]))
y_t_ema3 = np.dot(x, emapoints)where solve_weights is the function that solves the linear least square problem. You can find the implementation in ema_eq.py.
The result is the EMA with