The goal of multispat is to …
You can install the development version of multispat from GitHub with:
# install.packages("devtools")
devtools::install_github("clsong/multispat")This is a basic example which shows you how to solve a common problem:
suppressPackageStartupMessages(library(tidyverse))
library(multispat)
# set total number of species
gamma <- 3
# set species associations
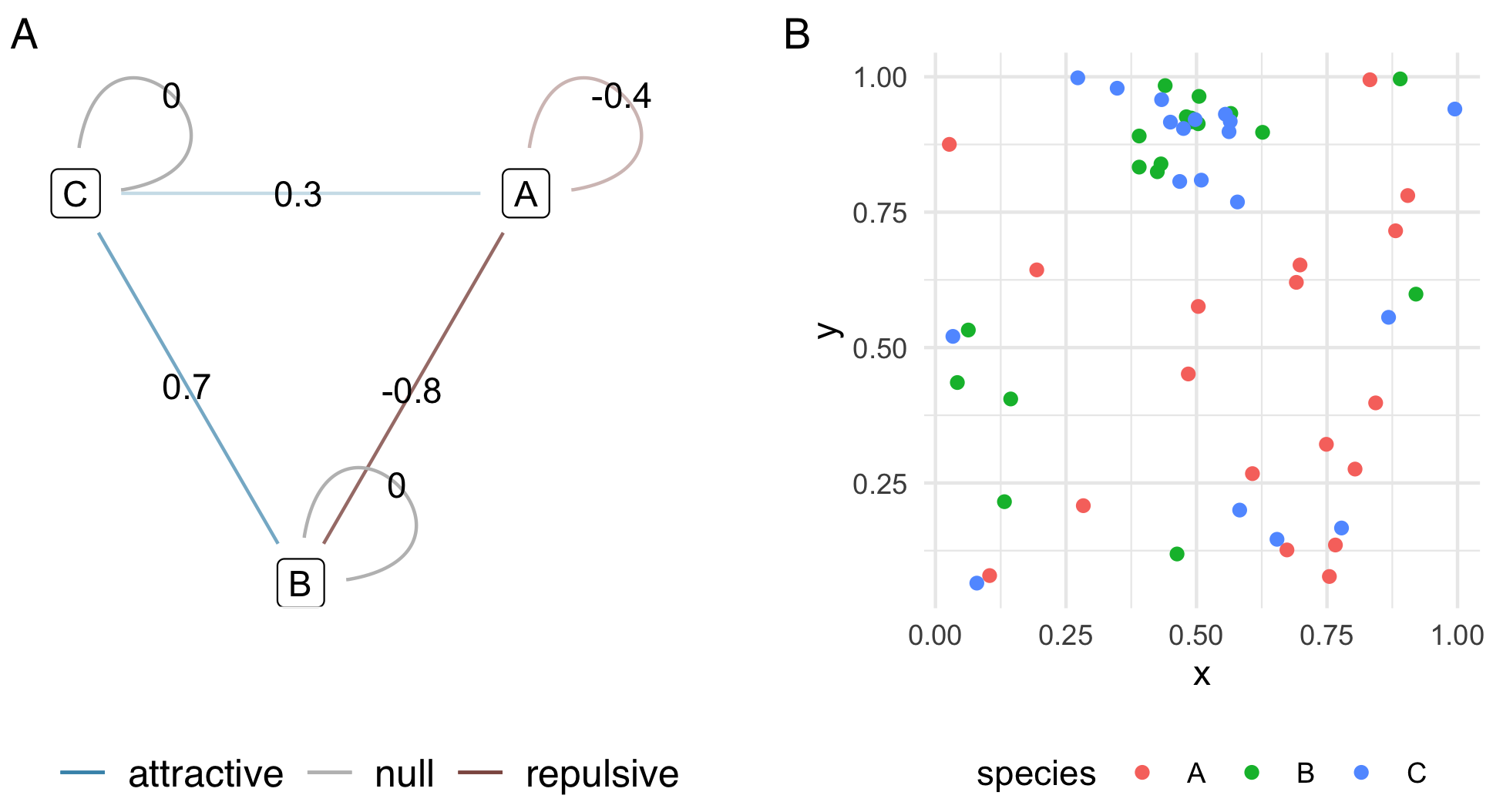
spatial_association <- matrix(NA, nrow = gamma, ncol = gamma)
diag(spatial_association) <- 0
spatial_association[1, 1] <- -.4
spatial_association[1, 2] <- -.8
spatial_association[1, 3] <- 0.3
spatial_association[2, 3] <- 0.7
spatial_association[lower.tri(spatial_association)] <- t(spatial_association)[lower.tri(spatial_association)]
# Visualize the spatial associations
p1 <- plot_spatial_association(spatial_association)
# Simulate the given point processes
df <- simulate_point_process(spatial_association, gamma, radius = .2)
# plot the spatial pattern
p2 <- df %>%
ggplot(aes(x, y, color = species)) +
geom_point() +
theme_minimal() +
theme(
aspect.ratio = 1,
legend.position = 'bottom'
)
library(patchwork)
p1 + p2 + plot_annotation(tag_levels = 'A')