Tools for Bayesian system identification.
bayesid delivers methods for System Identification using Bayesian neural networks with nonparametric noise processes [1]. Additionally, a autoregressive Bayesian neural network similar to [2] is implemented.
It is assumed that the user has or is able to install julia. This project has been developed in julia v1.7.1. The R package TimeSeries.OBeu is are also required for some functions used for plotting to work as expected.
The core files regarding the methods illustrated in [1] are located in src folder. Subfolders mcmc and models contain code for the Hamiltonian Monte Carlo sampler [*] and structs to implement neural networks using Flux, similar to neural networks implemented at Pkg DiffEqFlux.jl.
Scripts for simulating the logistic data as well as the real data used in the paper are located in data folder.
The folder examples contains simple scripts for running the sampler on data from data folder.
[*] hmc.jl is based on an implementation in SGMCMC.jl--see src/mcmc/LICENSE.
In order to install all dependencies in the current state, start julia from the location
bayesid is downloaded.
The project folder contains Manifest.toml and Project.toml files so you can type in the REPL
julia> ]
(@v1.7) pkg> activate .
(bayesid)> instantiateThis will make julia install all the necessary packages and dependencies for the available functions.
Having completed the installation process we can run an example by first loading necessary packages and files
using Plots, Random, Distributions, Flux, Zygote, LinearAlgebra
using StatsBase, StatsPlots, KernelDensity, DelimitedFiles, LaTeXStrings
using Parameters: @with_kw
include("../src/models/bnn.jl")
include("../src/mcmc/hmc.jl")
include("../src/utils.jl")
include("../src/npbnn.jl")
include("../src/arbnn.jl")
include("../src/plottools.jl")
include("../src/R/RUtils.jl")
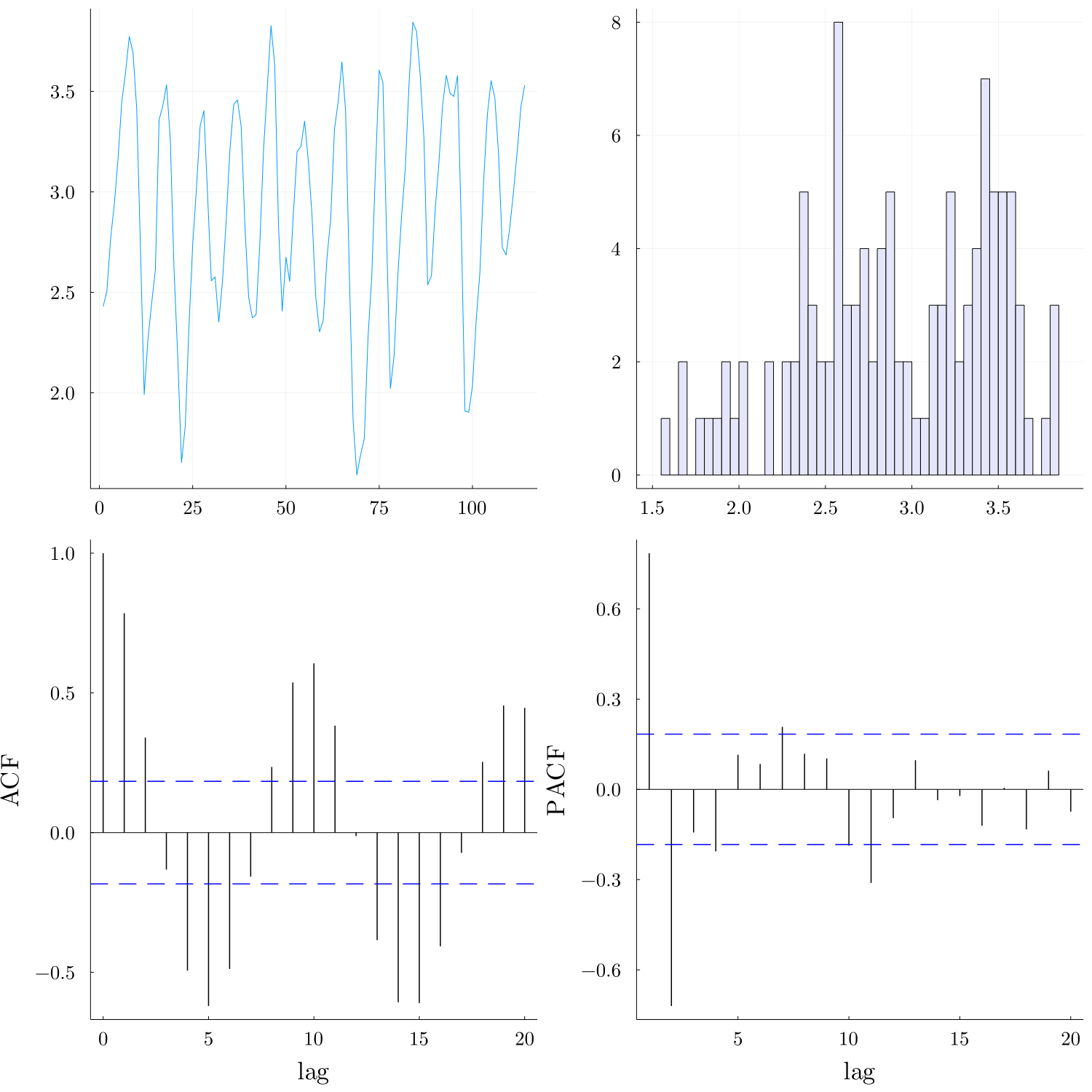
Next, we load the data and explore the time series along with the ACF and PACF.
data = log.(10, readdlm("./data/lynx.txt"));
data_plot = explore_data(data) # requires Rs TimeSeries.OBeu and tsThe PACF plot show significant partial autocorrelations at lags 1,2. Assuming that the data are coming from order 2 Markovian process, we construct the delayed time series
# split training data first 100 observations and generate the lagged time series via embed
lag = 2;
ytemp = data[1:end-14];
D = embed(ytemp, lag+1);
# train data
ytrain = convert(Array{Float64, 2}, hcat(D[:, 1]...));
xtrain = convert(Array{Float64, 2}, D[:, 2:end]');
ytest = data[101:end];Finally, a neural network is defined and a mutable struct containing the model and HMC parameters specification is defined. We identify the system using the npbnn function.
@with_kw mutable struct Args
net = g
maxiter = 40000 # maximum number of iterations
burnin = 20000 # burnin iterations
x = xtrain # lagged data
y = ytrain
geop = 0.5
hyper_taus = [1. 1. ;1. 1.]
ap = 1. # beta hyperparameter alpha for the geometric probability
bp = 1. # beta hyperparameter beta for the geometric probability
at = 0.05 # atoms gamma hyperparameter alpha
bt = 0.05 # atoms gamma hyperparameter beta
ataus = 5ones(2,2) # Gamma hyperprior on network weights precision
btaus = 5ones(2,2) # IG hyperprior on network weights precision
seed = 123
stepsize = 0.005
numsteps = 20
verb = 1000
npredict = 14
save = false
filename = "/sims/lynx/npbnn/"
end
@time est = npbnn();More details can be found in the /examples/canadianlynx.jl file.
We have included some state-of-the-art parametric models like ARMA based on R, using Julia's interoperability. These are in src/R/RUtils.jl
[1] Merkatas, C., & Särkkä, S. (2022). System identification using autoregressive Bayesian neural networks with nonparametric noise processes. Journal of Time Series Analysis.
[2] Nakada, Y., Matsumoto, T., Kurihara, T., & Yosui, K. (2005). Bayesian reconstructions and predictions of nonlinear dynamical systems via the hybrid Monte Carlo scheme. Signal processing, 85(1), 129-145.