Implementation for KDD'22 paper: GraphMAE: Self-Supervised Masked Graph Autoencoders.
We also have a Chinese blog about GraphMAE on Zhihu (知乎), and an English Blog on Medium.
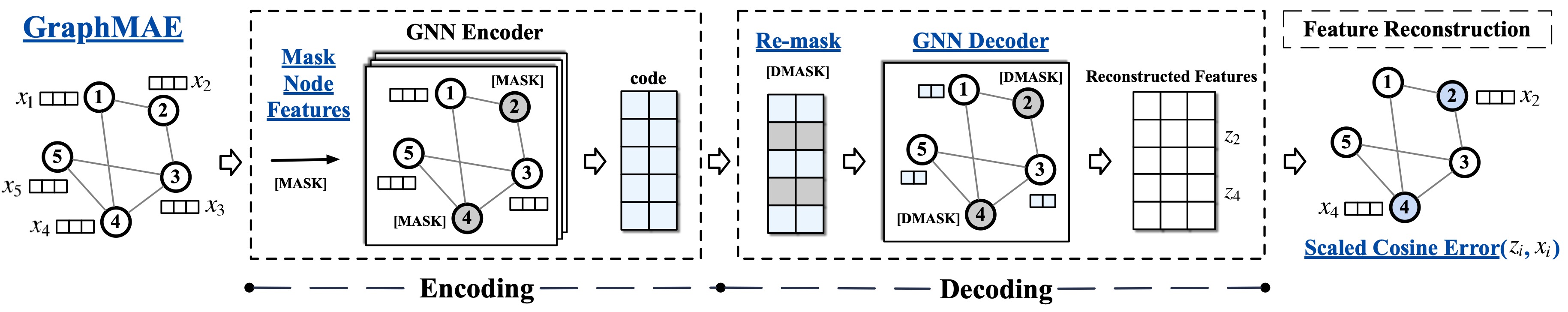
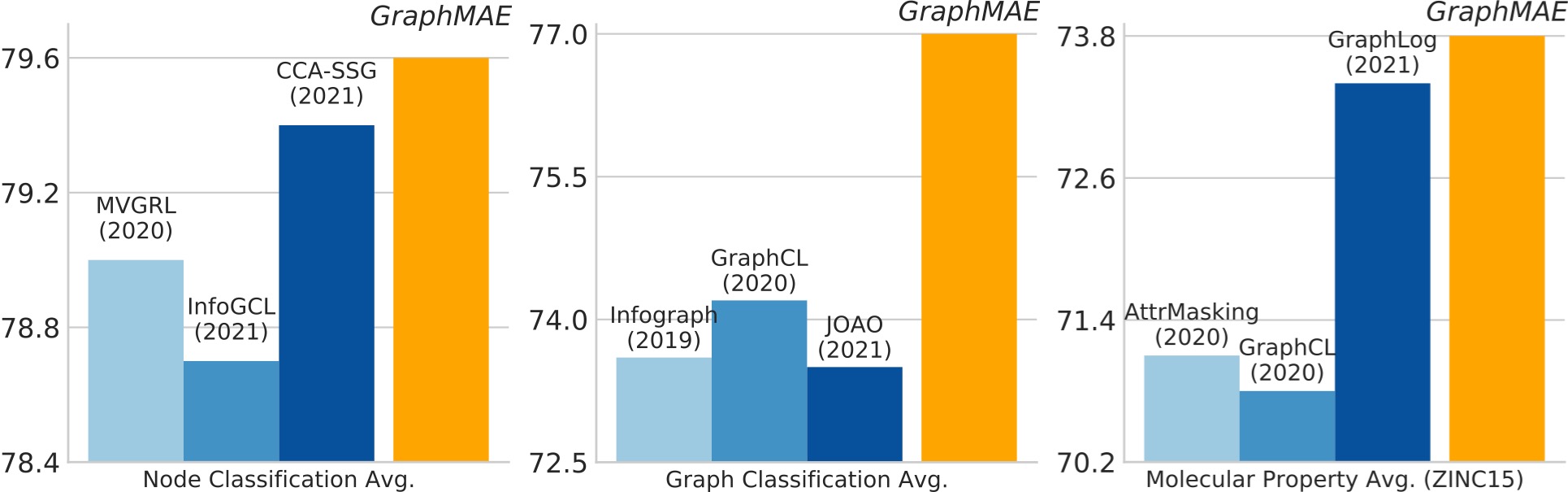
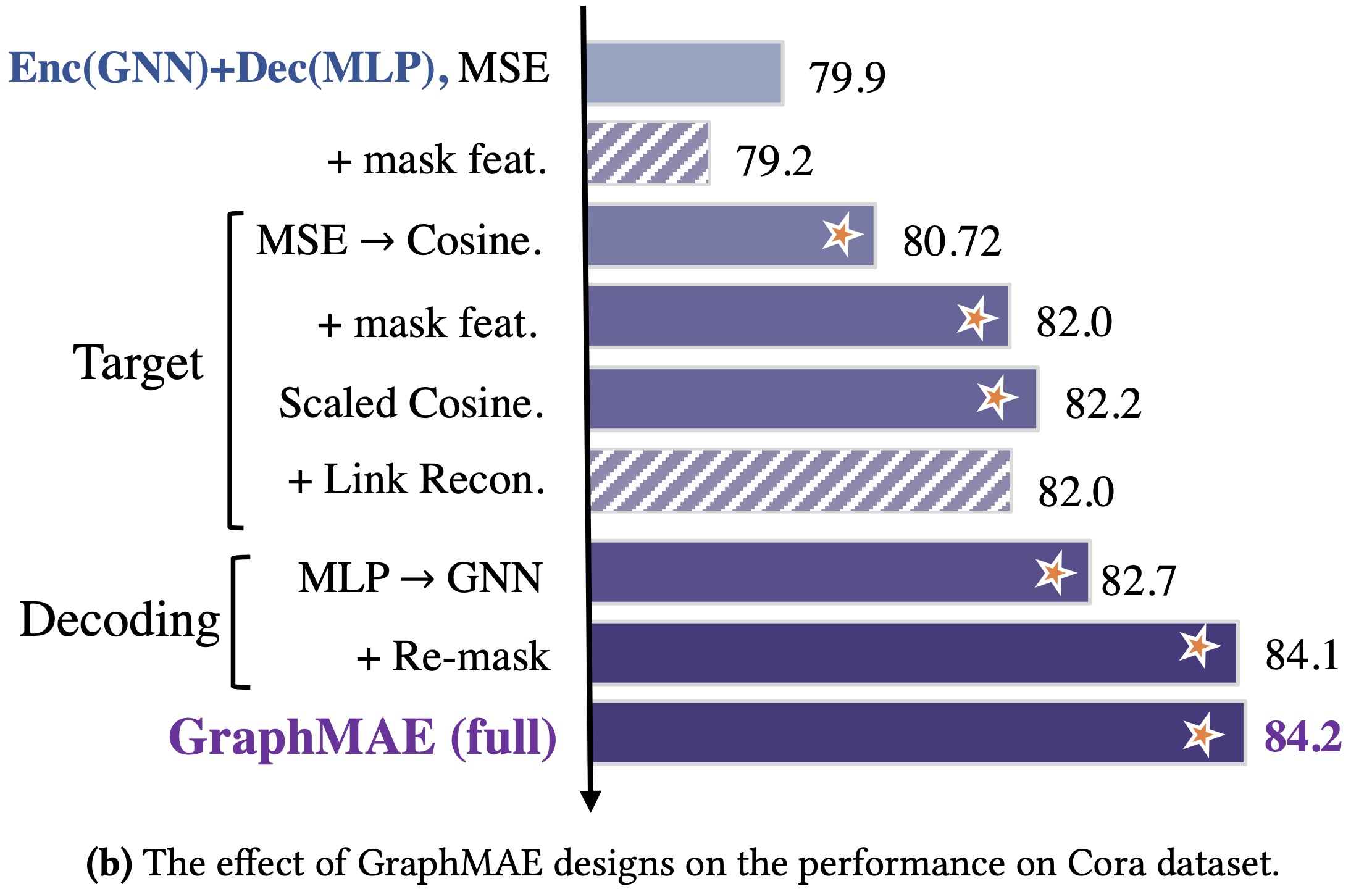
GraphMAE is a generative self-supervised graph learning method, which achieves competitive or better performance than existing contrastive methods on tasks including node classification, graph classification, and molecular property prediction.
[2022-12-14] The PYG implementation of GraphMAE for node / graph classification is available at this branch.
For quick start, you could run the scripts:
Node classification
sh scripts/run_transductive.sh <dataset_name> <gpu_id> # for transductive node classification
# example: sh scripts/run_transductive.sh cora/citeseer/pubmed/ogbn-arxiv 0
sh scripts/run_inductive.sh <dataset_name> <gpu_id> # for inductive node classification
# example: sh scripts/run_inductive.sh reddit/ppi 0
# Or you could run the code manually:
# for transductive node classification
python main_transductive.py --dataset cora --encoder gat --decoder gat --seed 0 --device 0
# for inductive node classification
python main_inductive.py --dataset ppi --encoder gat --decoder gat --seed 0 --device 0Supported datasets:
- transductive node classification:
cora,citeseer,pubmed,ogbn-arxiv - inductive node classification:
ppi,reddit
Run the scripts provided or add --use_cfg in command to reproduce the reported results.
Graph classification
sh scripts/run_graph.sh <dataset_name> <gpu_id>
# example: sh scripts/run_graph.sh mutag/imdb-b/imdb-m/proteins/... 0
# Or you could run the code manually:
python main_graph.py --dataset IMDB-BINARY --encoder gin --decoder gin --seed 0 --device 0Supported datasets:
IMDB-BINARY,IMDB-MULTI,PROTEINS,MUTAG,NCI1,REDDIT-BINERY,COLLAB
Run the scripts provided or add --use_cfg in command to reproduce the reported results.
Molecular Property Prediction
Please refer to codes in ./chem for molecular property prediction.
Datasets used in node classification and graph classification will be downloaded automatically from https://www.dgl.ai/ when running the code.
Node classification (Micro-F1, %):
| Cora | Citeseer | PubMed | Ogbn-arxiv | PPI | ||
|---|---|---|---|---|---|---|
| DGI | 82.3±0.6 | 71.8±0.7 | 76.8±0.6 | 70.34±0.16 | 63.80±0.20 | 94.0±0.10 |
| MVGRL | 83.5±0.4 | 73.3±0.5 | 80.1±0.7 | - | - | - |
| BGRL | 82.7±0.6 | 71.1±0.8 | 79.6±0.5 | 71.64±0.12 | 73.63±0.16 | 94.22±0.03 |
| CCA-SSG | 84.0±0.4 | 73.1±0.3 | 81.0±0.4 | 71.24±0.20 | 73.34±0.17 | 95.07±0.02 |
| GraphMAE(ours) | 84.2±0.4 | 73.4±0.4 | 81.1±0.4 | 71.75±0.17 | 74.50±0.29 | 96.01±0.08 |
Graph classification (Accuracy, %)
| IMDB-B | IMDB-M | PROTEINS | COLLAB | MUTAG | REDDIT-B | NCI1 | |
|---|---|---|---|---|---|---|---|
| InfoGraph | 73.03±0.87 | 49.69±0.53 | 74.44±0.31 | 70.65±1.13 | 89.01±1.13 | 82.50±1.42 | 76.20±1.06 |
| GraphCL | 71.14±0.44 | 48.58±0.67 | 74.39±0.45 | 71.36±1.15 | 86.80±1.34 | 89.53±0.84 | 77.87±0.41 |
| MVGRL | 74.20±0.70 | 51.20±0.50 | - | - | 89.70±1.10 | 84.50±0.60 | - |
| GraphMAE(ours) | 75.52±0.66 | 51.63±0.52 | 75.30±0.39 | 80.32±0.46 | 88.19±1.26 | 88.01±0.19 | 80.40±0.30 |
Transfer learning on molecular property prediction (ROC-AUC, %):
| BBBP | Tox21 | ToxCast | SIDER | ClinTox | MUV | HIV | BACE | Avg. | |
|---|---|---|---|---|---|---|---|---|---|
| AttrMasking | 64.3±2.8 | 76.7±0.4 | 64.2±0.5 | 61.0±0.7 | 71.8±4.1 | 74.7±1.4 | 77.2±1.1 | 79.3±1.6 | 71.1 |
| GraphCL | 69.7±0.7 | 73.9±0.7 | 62.4±0.6 | 60.5±0.9 | 76.0±2.7 | 69.8±2.7 | 78.5±1.2 | 75.4±1.4 | 70.8 |
| GraphLoG | 72.5±0.8 | 75.7±0.5 | 63.5±0.7 | 61.2±1.1 | 76.7±3.3 | 76.0±1.1 | 77.8±0.8 | 83.5±1.2 | 73.4 |
| GraphMAE(ours) | 72.0±0.6 | 75.5±0.6 | 64.1±0.3 | 60.3±1.1 | 82.3±1.2 | 76.3±2.4 | 77.2±1.0 | 83.1±0.9 | 73.8 |
If you find this work is helpful to your research, please consider citing our paper:
@article{hou2022graphmae,
title={GraphMAE: Self-Supervised Masked Graph Autoencoders},
author={Hou, Zhenyu and Liu, Xiao and Cen, Yukuo and Dong, Yuxiao and Yang, Hongxia and Wang, Chunjie and Tang, Jie},
journal={arXiv e-prints},
pages={arXiv--2205},
year={2022}
}