💡 This is the official implementation of the paper "MediCLIP: Adapting CLIP for Few-shot Medical Image Anomaly Detection"(MICCAI 2024) [arxiv].
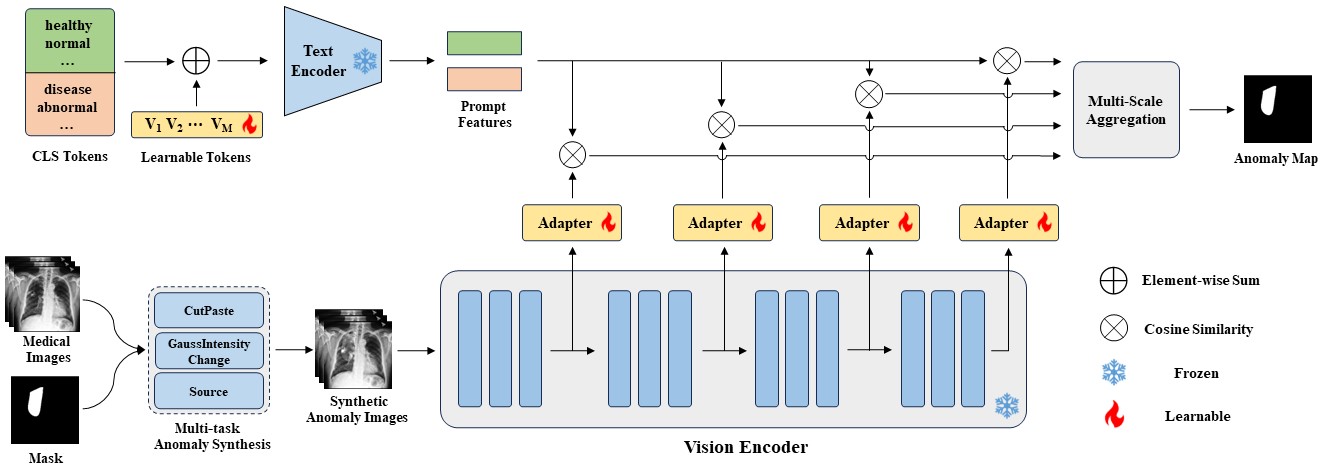
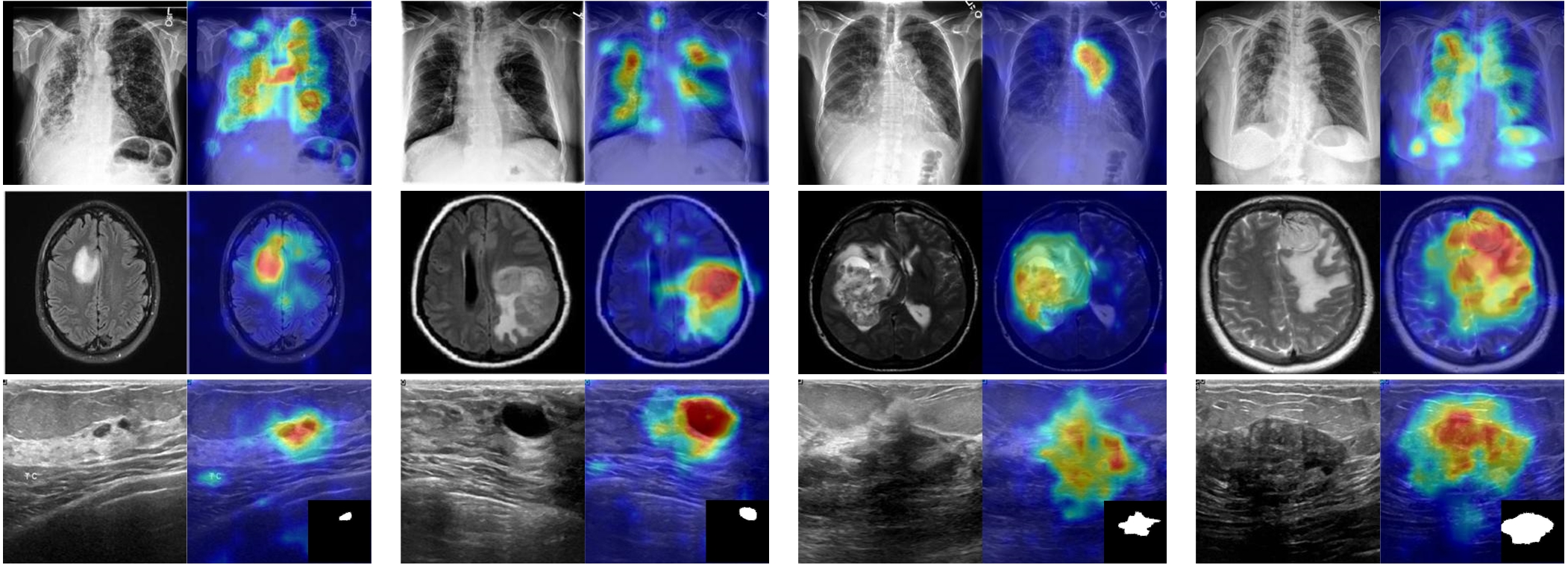
MediCLIP is an efficient few-shot medical image anomaly detection method, demonstrating SOTA anomaly detection performance with very few normal medical images. MediCLIP integrates learnable prompts, adapters, and realistic medical image anomaly synthesis tasks.
To run experiments, first clone the repository and install requirements.txt.
$ git clone https://github.com/cnulab/MediCLIP.git
$ cd MediCLIP
$ pip install -r requirements.txt
Download the following datasets:
- BUSI [Baidu Cloud (pwd8866)] [Google Drive] [Official Link]
- BrainMRI [Baidu Cloud (pwd8866)] [Google Drive] [Official Link]
- CheXpert [Baidu Cloud (pwd8866)] [Google Drive] [Official Link]
Unzip them to the data. Please refer to data/README.
To train the MediCLIP on the BrainMRI dataset with the support set size is 16:
$ python train.py --config_path config/brainmri.yaml --k_shot 16
To test the MediCLIP on the BrainMRI dataset:
$ python test.py --config_path config/brainmri.yaml --checkpoint_path xxx.pkl
Replace xxx.pkl with your checkpoint path.
Code reference: [CLIP] [CoOp] [Many-Tasks-Make-Light-Work].
If this work is helpful to you, please cite it as:
@inproceedings{zhang2024mediclip,
title={MediCLIP: Adapting CLIP for Few-shot Medical Image Anomaly Detection},
author={Ximiao Zhang, Min Xu, Dehui Qiu, Ruixin Yan, Ning Lang, and Xiuzhuang Zhou},
year={2024},
eprint={2405.11315},
archivePrefix={arXiv},
primaryClass={cs.CV}
}