Spatial mapping of human kidney by automated structural segmentation and registration across paired IHC, AF, and IMS datasets
Download QuPath for analysis of whole-slide IHC images. You will also need the Bioformats Extension.
QuPath scripts are found in qupath_dev/:
.groovyscripts run as QuPath macrosAutomate>Show Script Editor>File>Open...- open script, then
Run
.jsscripts are used in the ImageJ extension of QuPathExtensions>ImageJ>Export region to ImageJ- in ImageJ:
Plugins>Macros>Run
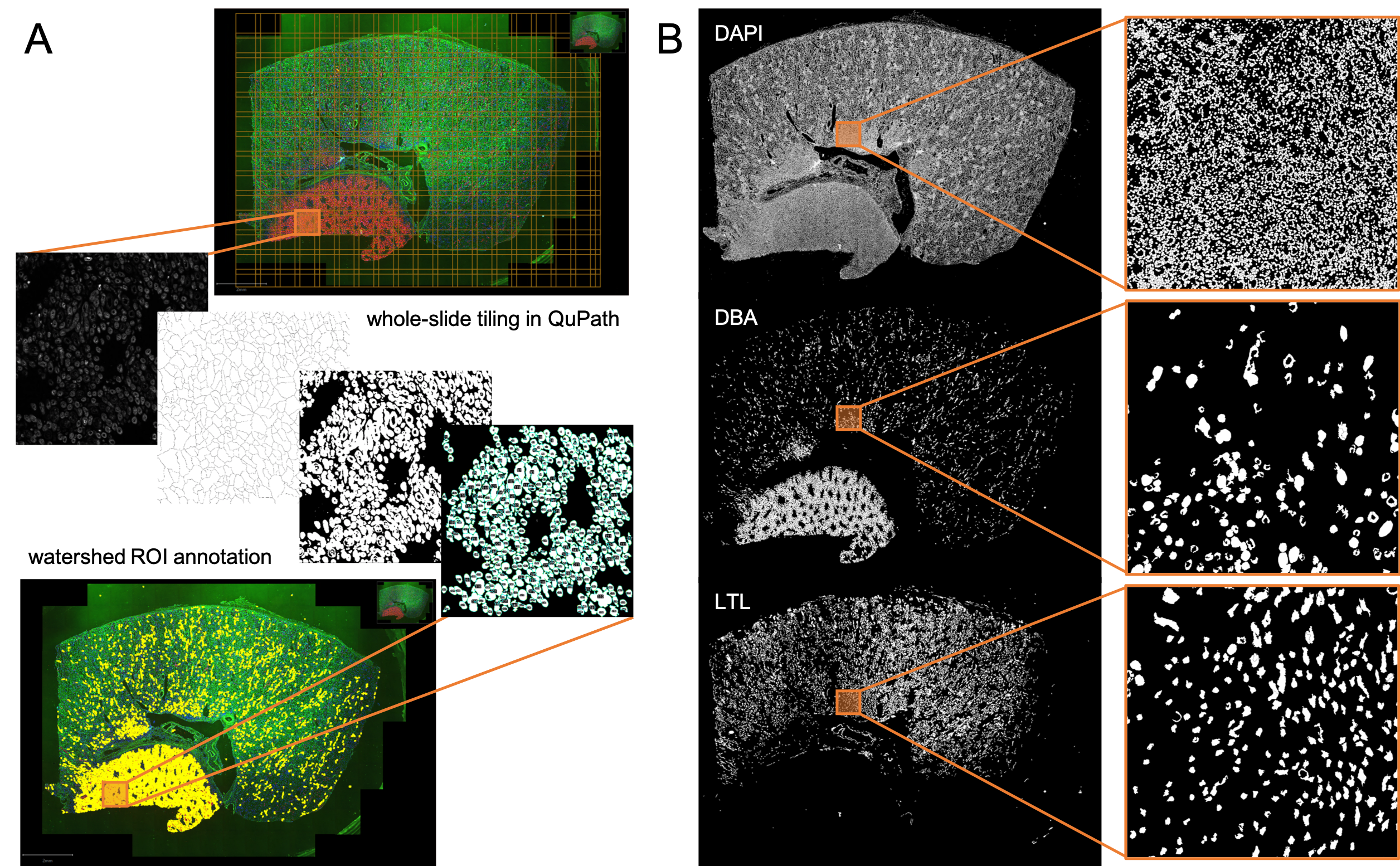
Figure 1. Workflow and example output for automated MxIF segmentation using QuPath. A) Whole-slide tiling workflow as implemented in javascript macro. Each tile of image is processed for watershed maxima and intensity threshold in a single channel; resulting ROIs returned as QuPath annotations. B) Binary image mask outputs in three MxIF channels, highlighting unique macro structures in the tissue.
pip install -r requirements.txt # make sure you have all necessary python packages