Unsupervised Pre-training Improves Tooth Segmentation in 3-Dimensional Intraoral Mesh Scans (Accept at MIDL2022)
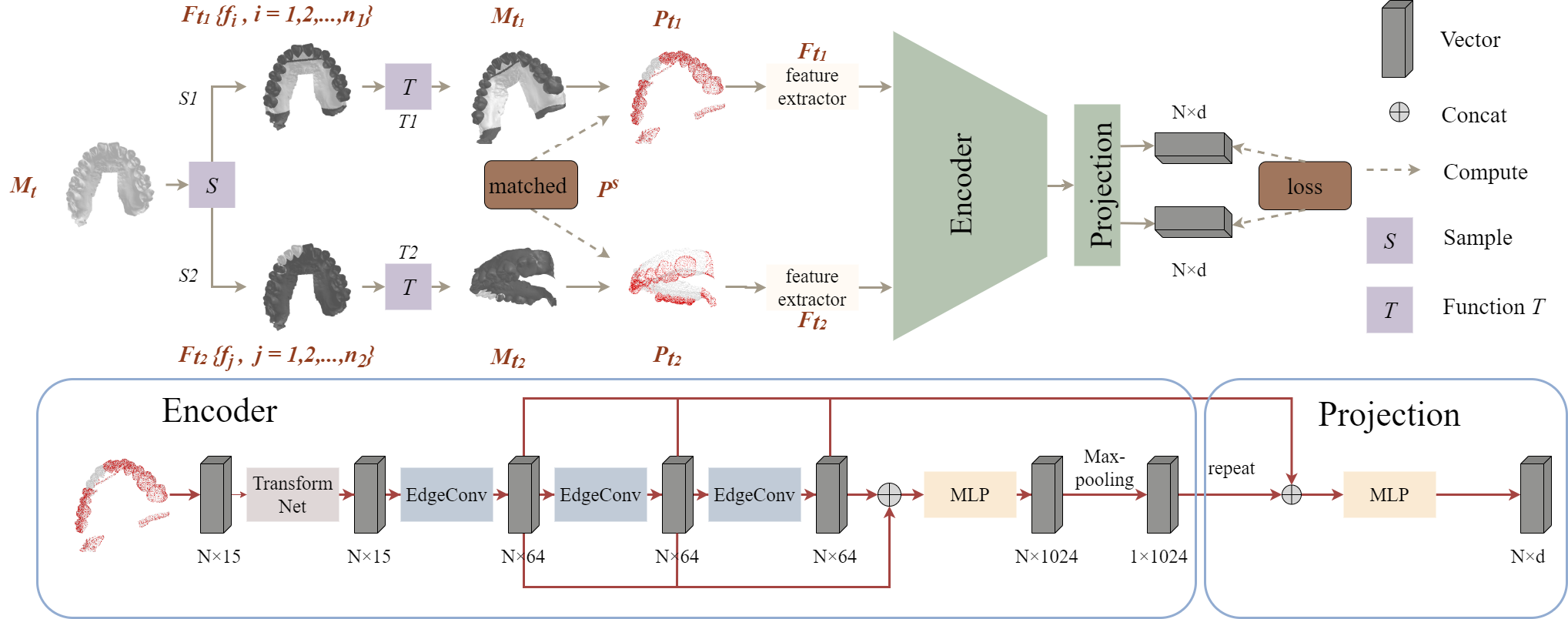
Accurate tooth segmentation in 3-Dimensional (3D) intraoral scanned (IOS) mesh data is an essential step for many practical dental applications. Recent research highlights the success of deep learning based methods for end-to-end 3D tooth segmentation, yet most of them are only trained or validated with a small dataset as annotating 3D IOS dental surfaces requires complex pipelines and intensive human efforts. In this paper, we propose a novel method to boost the performance of 3D tooth segmentation leveraging large-scale unlabeled IOS data. Our tooth segmentation network is first pre-trained with an unsupervised learning framework and point-wise contrastive learning loss on the large-scale unlabeled dataset and subsequently fine-tuned on a small labeled dataset. With the same amount of annotated samples, our method can achieve a mIoU of 89.38%, significantly outperforming the supervised counterpart. Moreover, our method can achieve better performance with only 40% of the annotated samples as compared to the fully supervised baselines. To the best of our knowledge, we present the first attempt of unsupervised pre-training for 3D tooth segmentation, demonstrating its strong potential in reducing human efforts for annotation and verification.
- Ubuntu 18.04
- CUDA 11.2
- Pytorch 1.9.0
- Open3d 0.9.0
- Trimesh 3.9.35
- MinkowskiEngine 0.5.4
For pre-training, put your dataset list to pretrain/dataset and rename it as overlap.txt.
dataset
├── 00000.stl
……
└── XXXXX.stl
The .stl is a simple 3-Dimensional Intraoral Mesh Scan without the label.
For fine-tuning, our codebase is based on AnTao97/dgcnn.pytorch. Similarly, please put your dataset lists to pretrain/dataset and rename them as teeth_train.txt,teeth_val.txt,teeth_test.txt.
├── trainset
│ ├── 00000.ply
│ ……
│ └── XXXXX.ply
├── valset
│ ├── 00000.ply
│ ……
│ └── XXXXX.ply
├── testset
│ ├── 00000.ply
│ ……
│ └── XXXXX.ply
We process the original .stl format and sample 20,000 surfaces for faster training. A set of preprocessing tools are provided in finetune/data_preprocess. Follow the README file in the finetune to generate the fine-tuning dataset. Here we list the details of .ply format data.
├── 00000.ply
│ ├── Face center[0]
│ ├── Face center[1]
│ ├── Face center[2]
│ ├── Shape Descriptor[0]
│ │ ……
│ ├── Shape Descriptor[8]
│ ├── Face normal[0]
│ ├── Face normal[1]
│ ├── Face normal[2]
│ └── label
Face center, shape descriptor and face normal have been defined in our paper.
CUDA_VISIBLE_DEVICES=0,1 python main.pyFor debugging purpose only, we provide a pretrained model in finetune/pretrained_weights/checkpoint.pth.
Fine-tuning:
CUDA_VISIBLE_DEVICES=0,1 python ddp_main.py --pretrain=./pretrained_weights/checkpoint.pth --lenient_weight_loading=True --exp_name=finetuneTrain from scratch:
CUDA_VISIBLE_DEVICES=0,1 python ddp_main.py --exp_name=from_scratchDuring fine-tuning, we only sample 20,000 points to train and test. If you want to obtain the labels of unsampled points, you can apply kNN strategy to the segmentation result. Meanwhile, there are also many methods to achieve it, e.g. putting all points into the model and graph-cut. So you can choose the appropriate method for post-processing with your needs.