Grab-N-Go Genomes: Automating Sequence Data Retrieval
GrabNGoGenomes was created with the "intro to biocomputing" student in mind. Often times, graduate students are new to bioinformatic skillsets and programs needed to perform their research. GrabNGoGenomes can help students get started by disentangling the sequence search and download process into a more streamlined process.
Information on how to configure GrabNGoGenomes can be obtained by forking GnGG_setup to your github repositories and following the directions below.
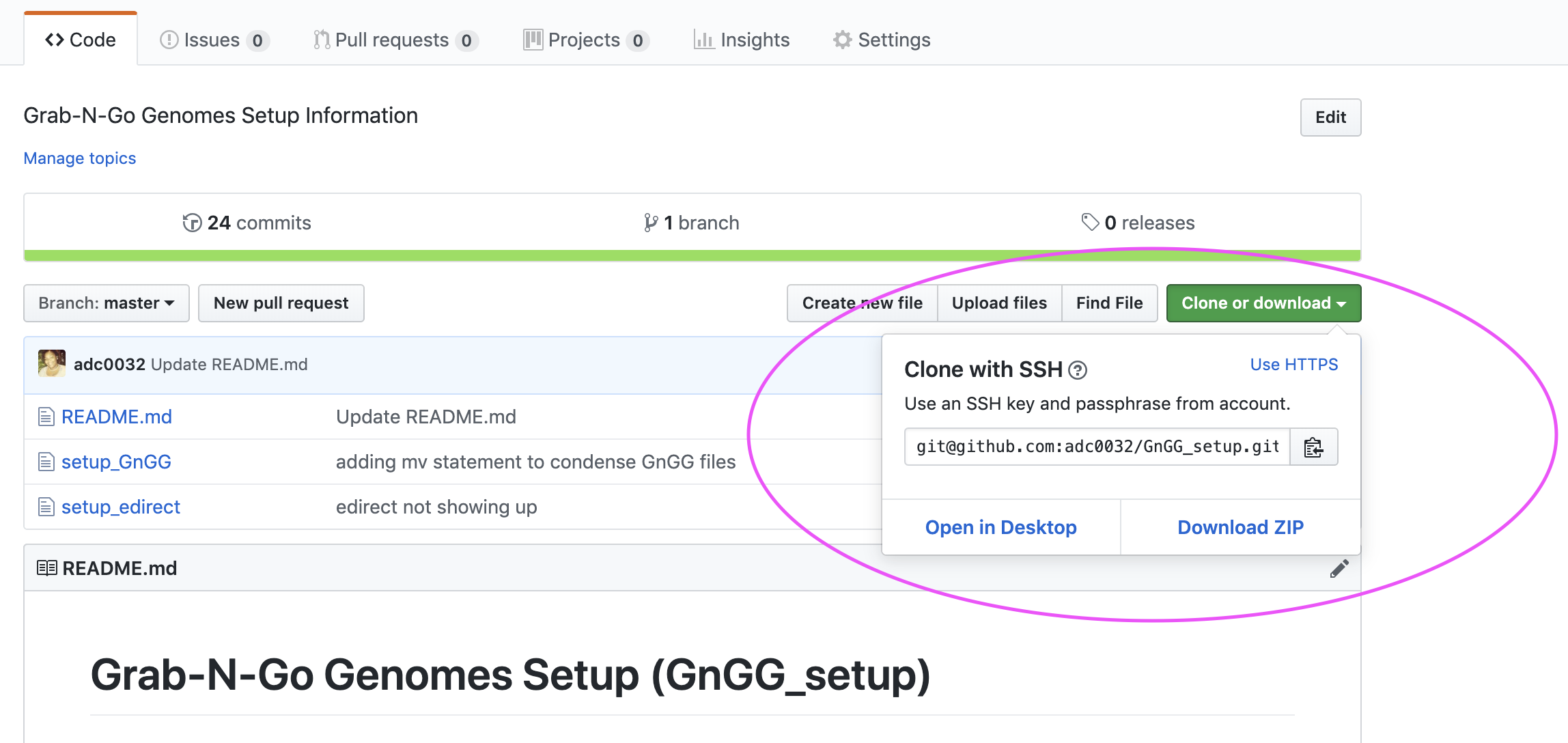
After forking the GnGG_setup repo, move to your forked version of the repo. From here, you will click clone to obtain the link for cloning the repository locally:
Run the following formatted code blocks and scripts in your terminal.
Cloning your forked GnGG_setup repository:
cd $HOME
git clone https://github.com/USER/GnGG_setup.gitUser should now have a directory titled GnGG_setup in /home/usr
Run:
cat GnGG_setup/setup_edirect
Copy output of previous command and paste to command line/terminal. The commands will execute. The last command ./edirect/setup.sh will populate the terminal, but requires user to press enter to execute.
User will see the following message once ./edirect/setup.sh is executed:
Trying to establish local installations of any missing Perl modules
(as logged in /home/usr/edirect/setup-deps.log).
Please be patient, as this step may take a little while.
Once the edirect setup script is completed, a success message is printed:
Entrez Direct has been successfully downloaded and installed.
In order to complete the configuration process, please execute the following:
echo "export PATH=\${PATH}:/home/usr/edirect" >> $HOME/.bashrc
or manually edit the PATH variable assignment in your .bash_profile file.
User should not execute the suggested command from the success output. Step is accounted for in the final GnGG setup script setup_GnGG
To complete configuration of GrabNGoGenomes, run:
sh GnGG_setup/setup_GnGG
If user attempts to execute this command before pasting output from preceding command cat /GnGG_setup/setup_edirect, the following error message is given:
Commands in ~/GnGG_setup/setup_edirect must be copied and pasted into your terminal before running this script
Once user pastes commands from the setup_edirect to the terminal, the following message is expect:
"Please be patient, as this step may take a little while. Entrez Direct has been successfully downloaded and installed..."
User does not need to run the output command given from pasting setup_edirect code to terminal, those steps are completed in setup_GnGG.
Successful execution prints finalization command:
GrabNGoGenomes Setup Complete!
Copy the following command into the terminal to finalize setup: export PATH=$PATH:$HOME/GrabNGoGenomes >& /dev/null || setenv PATH $PATH:$HOME/GrabNGoGenomes
Once complete, try running command get_SeqRec -h | get_SeqRec to get started
Run:
export PATH=$PATH:$HOME/GrabNGoGenomes >& /dev/null || setenv PATH $PATH:$HOME/GrabNGoGenomeThis will complete set up for GrabNGoGenomes. Run get_SeqRec -h or get_SeqRec to get started!
For more information about GrabNGoGenomes, visit Grab-N-Go Genomes