module load rclone
rclone copy --drive-shared-with-me "dtigoogledrive:/DR_cellranger test/" .Looking at DR-9163_25_S25_L001_R2_001.fastq.gz. See count_gRNAs.R for detail. Executed as:
Rscript scripts/count_gRNAs.RBriefly, the script
- reads in the sequence-containing rows from the
fastqfile - excludes reads with
Ns (there aren't many anyway) - finds reads with expected pattern of
'GTTG[ACTG]{20}GTTT'where the gRNA is precisely 20 nt sandwiched betweenGTTGandGTTT - matches exctracted gRNA sequence with those expected from
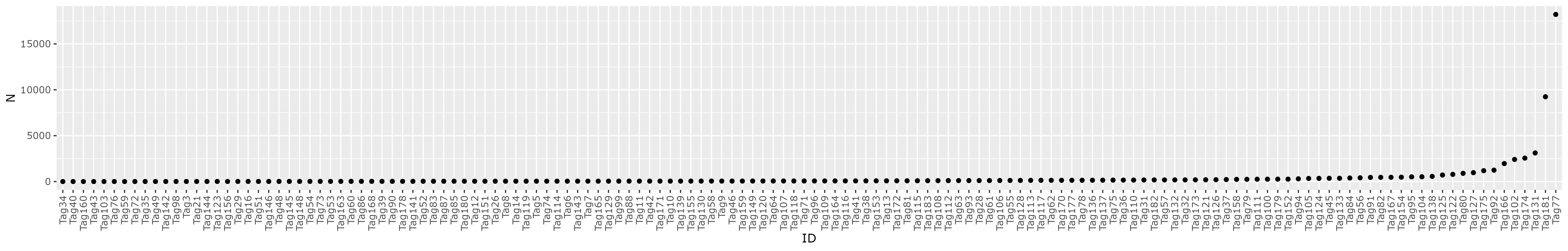
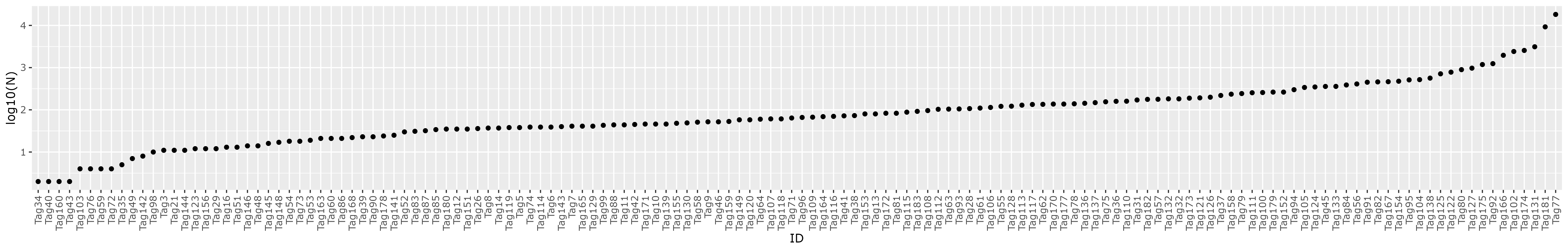
data/adt-tags-183sgRNAsequences-Tian2019.csv - plots distribution of guides with at least 1 occurence
See table: gRNA_distribution.tsv
cellranger.sh ran on Biowulf with sbatch. Right click and locally save the web summary here then open in your browser to view.