I am Veronica Reyes Galindo. This repository contains all the scripts of my Master's Thesis project at Instituto de Ecología, UNAM.
-
The online version of my Thesis is in UNAM website.
-
You can found "The paper" and other files about my project in my ResearchGate.
-
If you want to know more about the importance of the project to reforest the forest near to Mexico city, you can see a nice video and read a report in spanish by "Chilango" magazine.
And please send me a message in veronica.rg.pb@ gmail.com to clear all doubts about my Master's Thesis project.
 Photo by me: My Supervisor PhD. Alicia Mastretta-Yanes in middle of the "sacred fir cementery" in Santa Rosa Xochiac, 2017.
Photo by me: My Supervisor PhD. Alicia Mastretta-Yanes in middle of the "sacred fir cementery" in Santa Rosa Xochiac, 2017.
🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲🌲
The principal aim of my project was to : Assess if the tolerance of sacred fir (Abies religiosa) to exposure by O3, in a peripheral forest of the CDMX, is related to the genetic origin of the individuals or is given by differential expression of candidate genes.
To resolves my general aim, I have 3 particular aims. The analyzes to answer each aim can be found in separate directories with data and pictures:
-
Evaluate the differential expression of healthy and damaged trees in two periods of [O3] (TRANSCRIPTOMICS).
-
Identify origins of sacred fir with tolerance to O3 (GENOMICS).
-
Quantify the relative abundance of secondary metabolites in healthy y damaged trees in two periods of concentration of O3 (METABOLOMICS).
This is the Repository structure:
+----- Abies_religiosa_vs_ozone/
| +--README.md
| +--TRANSCRIPTOMICS/
| +--bin/
| +--data/
| +--metadata/
| +--outputs/
| +--README_transcriptomics.md
| +--GENOMICS/
| +--bin/
| +--data/
| +--metadata/
| +--outputs/
| +--README_genomics.md
| +--METABOLOMICS/
| +--bin/
| +--data/
| +--metadata/
| +--outputs/
| +--README_metabolomics.md
| +--INFO_PROJECT/
| +--Transcriptomic_analysis_about tropospheric_ozone_tolerance_in_Abies_religiosa_TB.pdf
| +--ABSTRACT-Transcriptomic_analysis_about_tropospheric_ozone_tolerance_in_Abies_religiosa.md
| +--README_info_project.md
| +--wonderful_images/
| +--all_images_in _this_repository.png
In addition, you can find in this repository:
📄 /README.md
README: is a intro about my project. This include the structure of my repository.
📁 /1.-GENOMICS: It is a genomic analysis from samples product of a GBS sequencing, ipyRAD was used to assemble de novo, VCFTools and plink to make more specific filters. The relationship was calculated, multiple SNPs were discarded in the same loci, a Mantel test, PCA and admixture were performed and the Heterocity was calculated.
📁 /2.-METABOLOMICS This is an analysis of metabolites measured with a gas chromatograph spectrum mass (GC-SM). Data from html files were loaded into tables that were subsequently calculated for their relative abundance. The values between samples were compared from a barplot, ANOVA and finally a PCA analysis was made with the final metabolites.
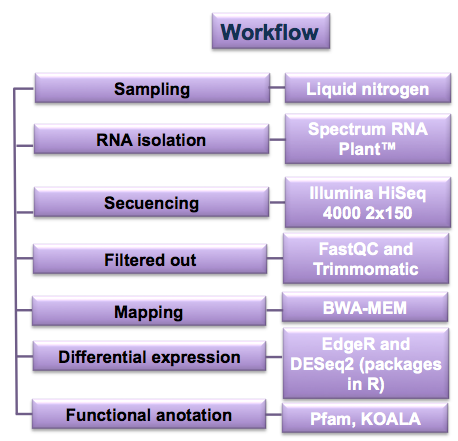
📁 /3.-TRANSCRIPTOMICS This is a transcriptomic analysis from samples sequenced with RNAseq. Samples were cut(Timmomatic) and mapped (BWA) to a reference transcriptome. Sequence counting was carried out through command lines in Rstudio that subsequently allowed the evaluation of differential expression between samples. From the counting table, a volcanoplot was performed to exemplify the overexpressed and underexpressed genes.
📁 /4.-INFO_PROJECT: Here are exhibitions, summaries and analysis of the final data of this repository.
📁 /5.-wonderful_images: This directory stores images that help complement and exemplify my README files and my issues. NOTE: The product images of R scripts are in the corresponding "outputs" directory for each analysis (GENOMICS, METABOLOMICS and TRANSCRIPTOMICS).
The principal analysis are:
Check more info about this pipeline in the README_genomics.
To see a short summary about the analysis of the final data you can go to Analysis_genomics
Quantify the relative abundance of secondary metabolites in healthy y damaged trees in two periods of concentration of O3
Check more info about this pipeline in the README_metabolomics.
To see a short summary about the analysis of the final data you can go to Analysis_metabolomics
Check more info about this pipeline in the README_transcriptomics.
To see a short summary about the analysis of the final data you can go to Analysis_transcriptomics