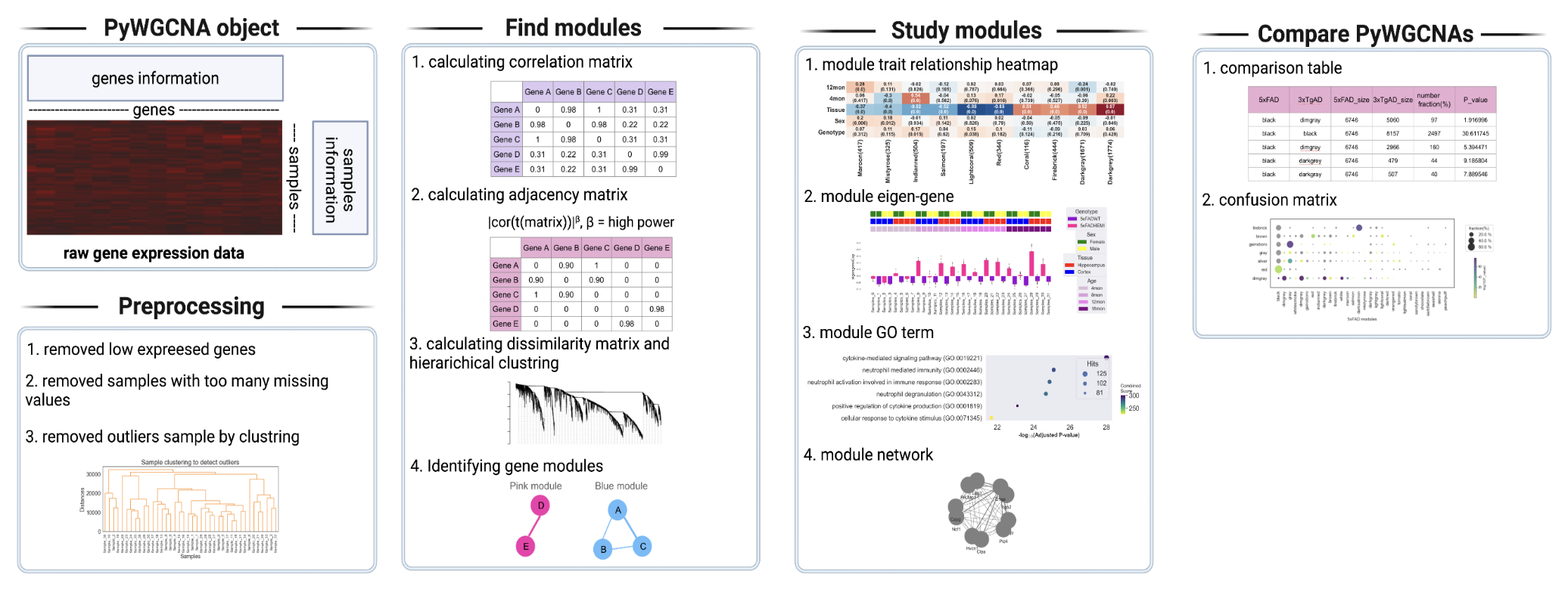
PyWGCNA is a Python library designed to do Weighted correlation network analysis (WGCNA) can be used for finding clusters (modules) of highly correlated genes, for summarizing such clusters using the module eigengene for relating modules to one another and to external sample traits (using eigengene network methodology), and for calculating module membership measures. Users can also compare WGCNA from different datasets including single cell gene markers.
PyWGCNA's full documentation can be found at here
To install PyWGCNA, python version 3.7 or greater is required.
Install the most recent release, run
pip install PyWGCNA
git cloning the PyWGCNA repository, going to the PyWGCNA directory, run
pip install .
- Data input, cleaning and pre-processing: How data format look like
- Quick Start: How to load data into PyWGCNA and find modules and analyse them
- Compare two PyWGCNA objects: How to compare two PyWGCNA objects
- Compare PyWGCNA objects to gene marker list: How to compare PyWGCNA objects to gene marker from single cell data
- Functional enrichment analysis : How to perform functional enrichment analysis including GO, KEGG and REACTOME in PyWGCNA object
- Visualize modules as network: How to visualize PyWGCNA objects as a network
- Recover Protein-Protein Interaction: How to find and plot PPI using STRING database.
If you are unfamiliar with R refrence WGCNA, we suggest reading the original WGCNA publication:
- WGCNA: an R package for weighted correlation network analysis
- Describe various features of the WGCNA package for the R programming language
Please cite our paper when using PyWGCNA:
Rezaie, Narges, Fairlie Reese, and Ali Mortazavi. "PyWGCNA: A Python package for weighted gene co-expression network analysis." bioRxiv (2022). https://www.biorxiv.org/content/10.1101/2022.08.22.504852v1.abstract