PDBClean
PDBClean offers curation tools for structural ensemble deposited in the Protein Data Bank.
For installation instructions, please see below.
Tutorials
Downloading and curating a structural ensemble
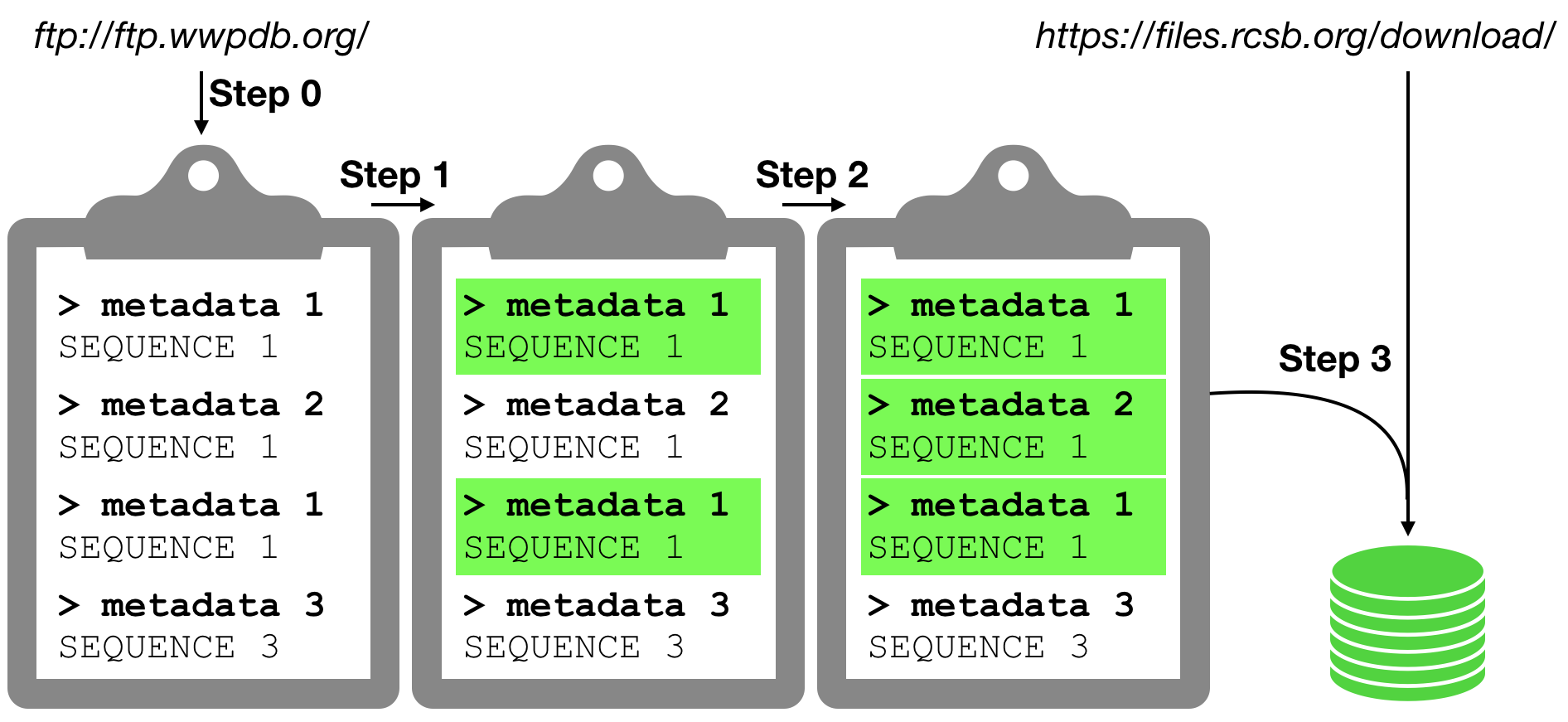
The overall protocol is broken down in elementary sequential steps described in the following notebooks
0. Download a structural ensemble from the RCSB PDB
1. Cleaning the CIF files just downloaded
2. Assign MolID to the entities found in the CIF files
Sharing curated dataset
We provide some ways to upload datasets to and download datasets from OSF, together with our examples.
Pulling and pushing datasets on OSF
List of datasets curated by the Levitt Lab
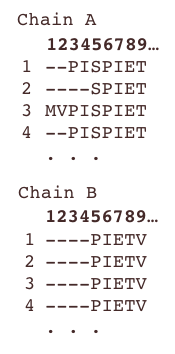
Chain and atom selection
Extracting a homogeneous dataset
For many types of analysis, one would need to be able to load the dataset as a feature-by-sample array that requires all samples to exhibit the same features. This homogeneization step is not unique
Extracting a homogeneous dataset
Analysis of the resulting dataset
Conformational heterogeneity
Installation
Download from Pypi
For now we only uploaded the package to TestPypi, so you also need to install the required tools listed below:
pip install --index-url https://test.pypi.org/simple/ --no-deps PDBClean
Download from Github
Assuming you have the required tools and libraries listed below, just type:
git clone https://github.com/csblab/PDBClean.git
python setup.py install