██████ ▓█████ ███▄ ▄███▓ ▄▄▄
▒██ ▒ ▓█ ▀ ▓██▒▀█▀ ██▒▒████▄
░ ▓██▄ ▒███ ▓██ ▓██░▒██ ▀█▄
▒ ██▒▒▓█ ▄ ▒██ ▒██ ░██▄▄▄▄██
▒██████▒▒░▒████▒▒██▒ ░██▒ ▓█ ▓██▒
▒ ▒▓▒ ▒ ░░░ ▒░ ░░ ▒░ ░ ░ ▒▒ ▓▒█░
░ ░▒ ░ ░ ░ ░ ░░ ░ ░ ▒ ▒▒ ░
░ ░ ░ ░ ░ ░ ░ ▒
░ ░ ░ ░ ░ ░
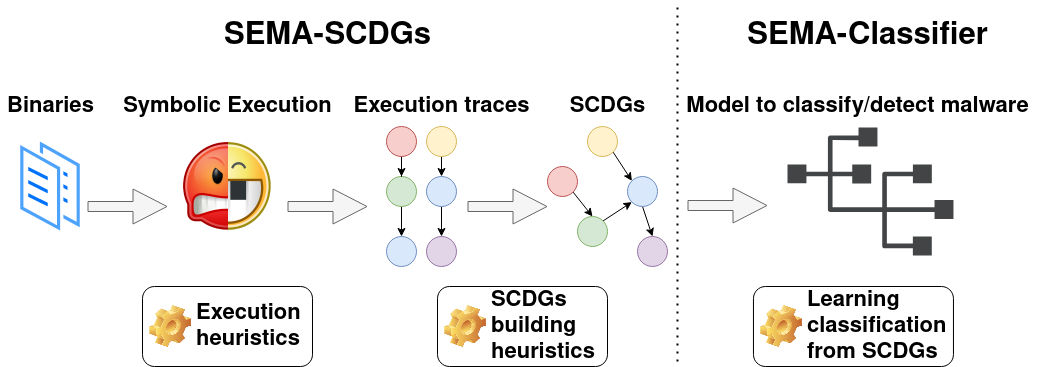
Our toolchain is represented in the following figure and works as follows:
- A collection of labelled binaries from different malware families is collected and used as the input of the toolchain.
- Angr, a framework for symbolic execution, is used to execute binaries symbolically and extract execution traces. For this purpose, different heuristics have been developed to optimize symbolic execution.
- Several execution traces (i.e., API calls used and their arguments) corresponding to one binary are extracted with Angr and gathered together using several graph heuristics to construct a SCDG.
- These resulting SCDGs are then used as input to graph mining to extract common graphs between SCDGs of the same family and create a signature.
- Finally, when a new sample has to be classified, its SCDG is built and compared with SCDGs of known families using a simple similarity metric.
This repository contains a first version of a SCDG extractor. During the symbolic analysis of a binary, all system calls and their arguments found are recorded. After some stop conditions for symbolic analysis, a graph is built as follows: Nodes are system calls recorded, edges show that some arguments are shared between calls.
When a new sample has to be evaluated, its SCDG is first built as described previously. Then, gspan is applied to extract the biggest common subgraph and a similarity score is evaluated to decide if the graph is considered as part of the family or not. The similarity score S between graph G' and G'' is computed as follows:
Since G'' is a subgraph of G', this is calculating how much G' appears in G''.
Another classifier we use is the Support Vector Machine (SVM) with INRIA graph kernel or the Weisfeiler-Lehman extension graph kernel.
A web application is available and is called SemaWebApp. It allows to manage the launch of experiments on SemaSCDG and/or SemaClassifier.
-
A Makefile is provided to ease the usage of the toolchain, run
make helpfor more information about the available commands
Main authors of the projects:
-
Charles-Henry Bertrand Van Ouytsel (UCLouvain)
-
Christophe Crochet (UCLouvain)
-
Khanh Huu The Dam (UCLouvain)
-
Oreins Manon (UCLouvain)
Under the supervision and with the support of Fabrizio Biondi (Avast)
Under the supervision and with the support of our professor Axel Legay (UCLouvain) (:heart:)