A tool to assign VAginaL DYnamic categories to individuals based on their VALENCIA CST assignments.
If using any part of this work, please cite:
Hugerth, L.W., Krog, M.C., Vomstein, K. et al. Defining Vaginal Community Dynamics: daily microbiome transitions, the role of menstruation, bacteriophages, and bacterial genes. Microbiome 12, 153 (2024). https://doi.org/10.1186/s40168-024-01870-5
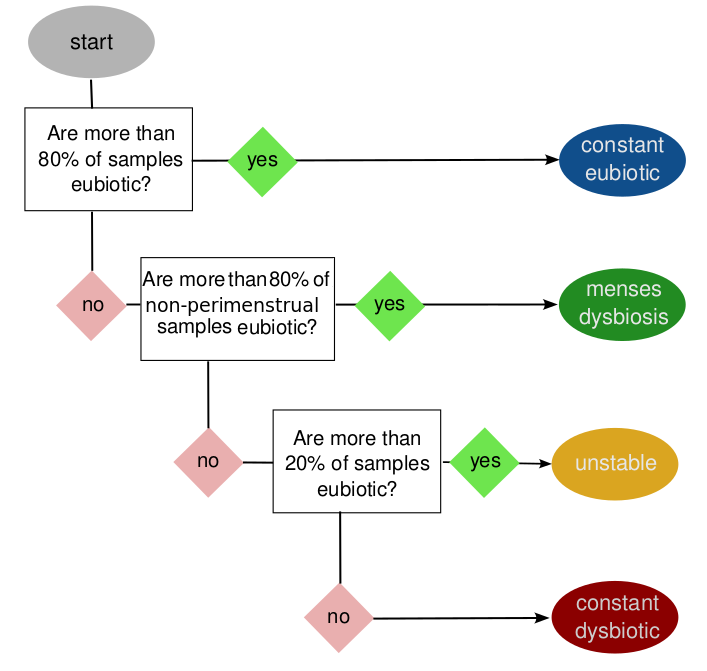
Vaginal microbiomes are tightly linked to the menstrual cycle, through menstrual bleeding and/or changes to the mucosal lining associated with estrogen levels. These microbiomes can also be affected by other factors, such as intercourse, hygiene, medication and stochastic processes. VALODY is designed to classify vaginal time-series according to the frequency and temporal pattern of their community state types in relation to subjects' menstrual cycles:
The user must therefore define:
- which CST are considered eubiotic (by default: I, II and V)
- Which days of the cycle are affected by menses.
Empirically, we find that the last 2-3 days of the luteal phase should be included in the "perimenstrual" period, so that in a 28 day cycle, days 1-8 as well as 26-28 are considered perimenstrual and days 9-25 as "central".
The inputs to valody are the output of VALENCIA as well as a comma-separated file with the format
| sampleID | subjectID | menses |
|---|---|---|
| sample1 | subj1 | 1 |
| sample2 | subj1 | 1 |
| ... | ... | ... |
| sampleN | subjM | 0 |
Where sample IDs should be identical to the ones in the VALENCIA output file
and menses should be marked as 1 for perimenstrual samples and 0 otherwise.
VALODY can then be run as:
./valody.py -i INPUT -m METADATA -o OUTPUT [-d DYSBIOSIS] [-e EUBIOSIS] [-s]
-i INPUT, --input INPUT, path to VALENCIA output
-m METADATA, --metadata METADATA, CSV file with 'sampleID,subjectID,menses', where menses takes 1 for yes and 0 for no
-o OUTPUT, --output OUTPUT, Output csv file
-d DYSBIOSIS, --dysbiosis DYSBIOSIS, comma-separated list of CST or sub-CST considered dysbiotic. Default: I, II, V
-e EUBIOSIS, --eubiosis EUBIOSIS, comma-separated list of CST or sub-CST considered eubiotic. Default: III, IV
-s, --subtypes, optional: use CST subtypes instead of main types; requires eubiosis and dysbiosis argument