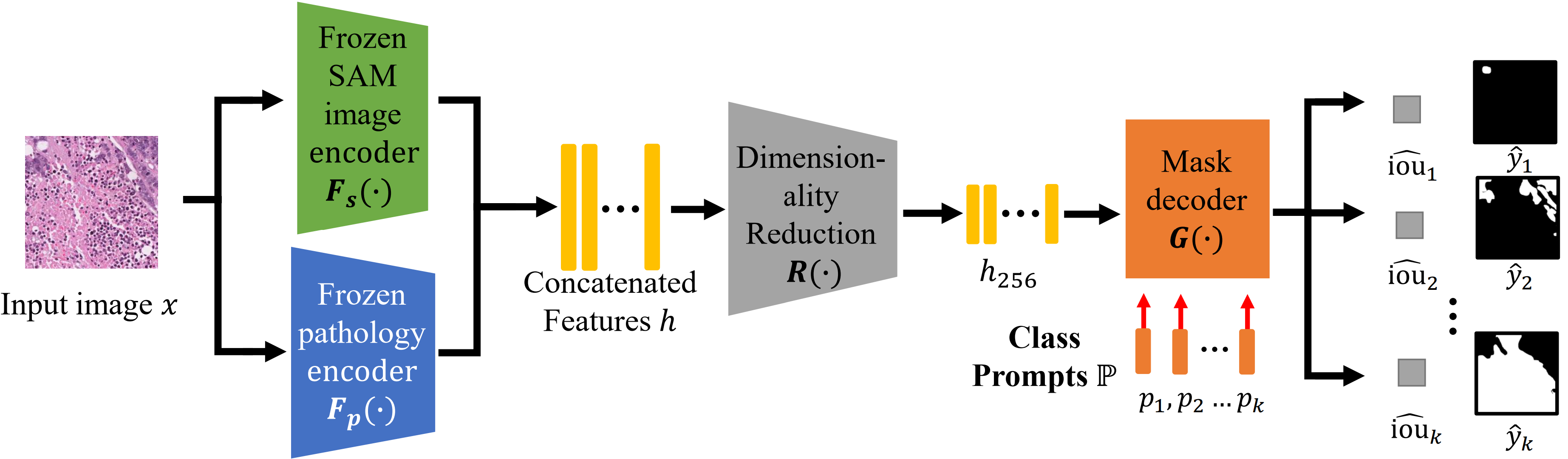
Pytorch implementation for the SAM-PAth framework described in the paper SAM-Path: A Segment Anything Model for Semantic Segmentation in Digital Pathology, arxiv and (MedAGI 2023, accepted for oral presentation).
Install Anaconda/miniconda.
Install the dependencies of SAM. Please do not install the original SAM itself as we made some modifications.
Then Install required packages:
$ pip install monai torchmetrics==0.11.4 pytorch_lightning==2.0.2 albumentations box wandb
Our dataset is organized as csv indicated datasets. All the images and masks should be stored in a directory and the path of this directory (dataset_root) should be set in the config file.
The root directory should contain two sub-directories img and mask. All the input images and masks should be directly put into these two sub-directories respectively.
Our preprocessed dataset can be downloaded from: https://drive.google.com/drive/folders/1BUPZz3nB52J5zRs1ZcEvNK03zw18BeLN?usp=sharing
The file names and train/validation/test separation are listed in the csv file. This csv file should contain 2 columns: img_id and fold. img_id is the filename of input image without the file extension. fold is the integer label of an input image. -1 means it is a test sample. We use the fold=0 as the validation dataset and fold=1,2,3,4 as the training dataset. The csv files we used are provided in the dataset_cfg folder.
We used train.py to train and evaluate our framework.
usage: main.py [--config CONFIG_PATH] [--devices GPU_ID]
[--project PROJECT_NAME] [----name RUN_NAME]
For example:
python main.py --config configs.BCSS --devices 0 --project sampath --name bcss_run0
python main.py --config configs.CRAG --devices 1 --project sampath --name crag_run0
Config files are located in the configs folder. Not the extension .py should not be included and the sub-folders should be linked by .
Pretrained SAM and HIPT models can be downloaded from their ogriginal repository: SAM and HIPT.
We used predict.py to predict the mask of a dataset.
usage: predict.py [--config CONFIG_PATH] [--devices GPU_ID]
[--pretrained path_to_pretrained_weights]
[--input_dir path_to_image_directory]
[--data_ext image_extension]
[--output_dir path_to_output_directory]
For example:
python predict.py --config configs.BCSS --input_dir path_to_image_directory --data_ext .png --output_dir path_to_output_directory --pretrained /.../model.ckpt --devices 2
Note that we always use label 0 as the unlabeled region. If the dataset does not contain any unlabeled region, all the predicted masks need be subtracted by 1.
If you have any questions or concerns, feel free to report issues or directly contact us (Jingwei Zhang jingwezhang@cs.stonybrook.edu).
If you use the code or results in your research, please use the following BibTeX entry.
@article{zhang2023sam,
title={SAM-Path: A Segment Anything Model for Semantic Segmentation in Digital Pathology},
author={Zhang, Jingwei and Ma, Ke and Kapse, Saarthak and Saltz, Joel and Vakalopoulou, Maria and Prasanna, Prateek and Samaras, Dimitris},
journal={arXiv preprint arXiv:2307.09570},
year={2023}
}