Codes of our ICLR'24 paper. [Paper Link], [Website], [Demo]
Authors: Sihang Li*, Zhiyuan Liu*, Yanchen Luo, Xiang Wang†, Xiangnan He†, Kenji Kawaguchi, Tat-Seng Chua, Qi Tian
* Equal Contribution
† Corresponding
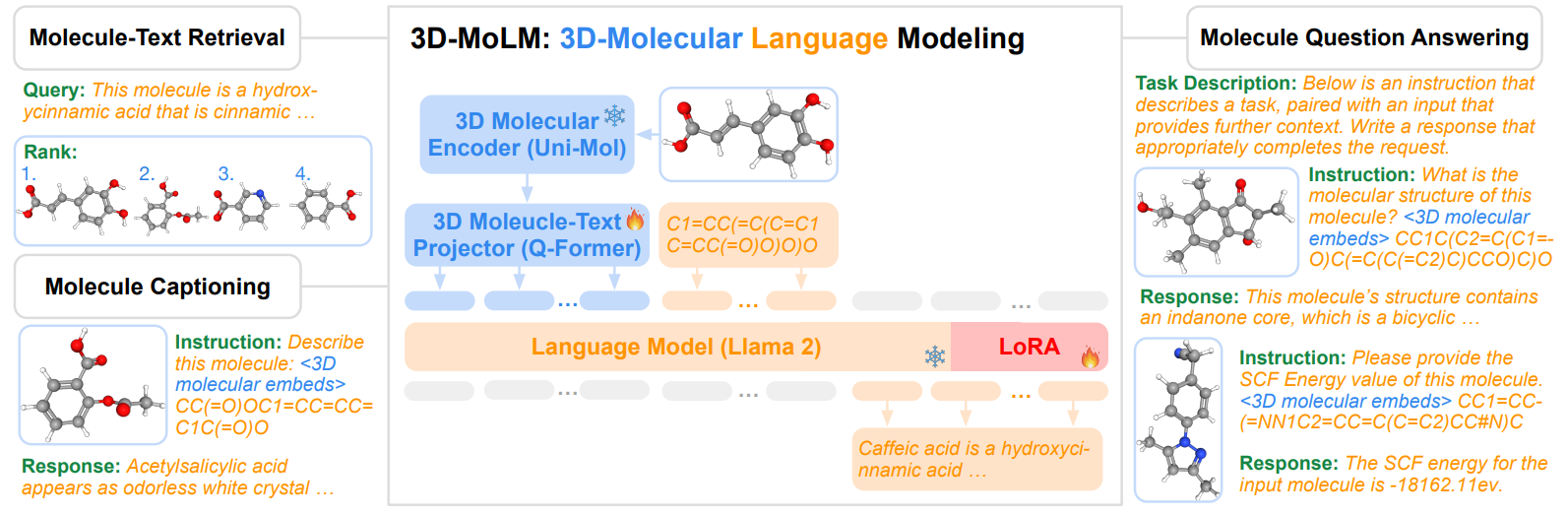
- 3D-MoLM is a versatile molecular LM that can be applied for molecule-text retrieval, molecule captioning, and molecule question-answering tasks.
- 3D-MoLM employs a 3D molecule-text projector to bridge the modality gap between a 3D molecular encoder and an LM, enabling the LM to perceive 3D molecular structures.
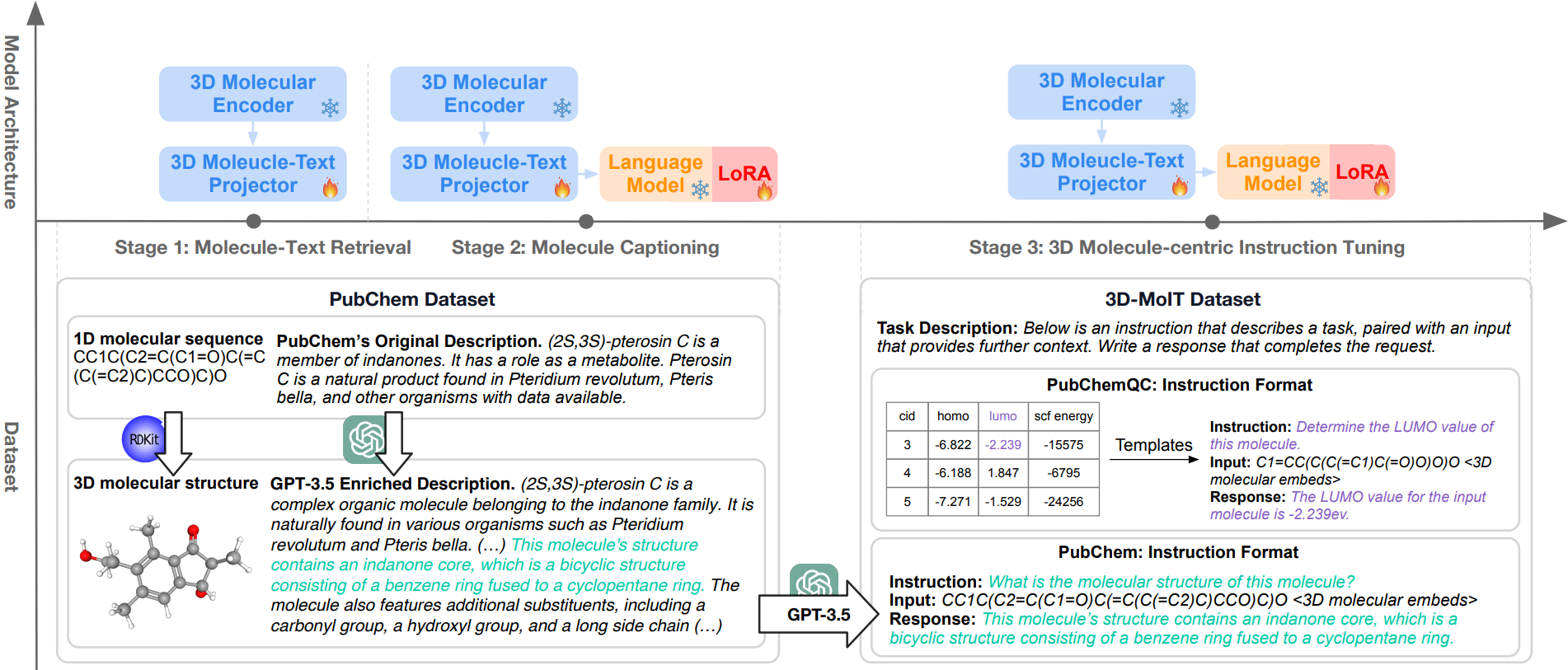
To tackle the two challenges of 3D molecule-text alignment and 3D molecule-centric instruction tuning, we delineate a three-stage training pipeline for 3D-MoLM, including 1) 3D molecule-text representation learning, 2) 3D molecule-text alignment via text generation, and 3) instruction-based fine-tuning.
3D Molecule-Text Alignment maps 3D molecular representations into the input textual space where the LM can understand.
- Data Preparation – PubChem. We collect molecular SMILES-text pairs from PubChem, and further employ GPT-3.5 to enrich the less annotated molecular descriptions. Molecular 3D conformations are obtained by running the MMFF algorithm in RDKit. As a result, 316K 3D molecule-text pairs are obtained for the alignment pretraining and downstream tasks of molecule-text retrieval and molecule captioning.
- Stage 1: 3D Molecule-Text Representation Learning cultivates Q-Former’s ability to extract molecular features that resonate profoundly with the corresponding text
- Stage 2: 3D Molecule-Text Alignment via Generative Learning connect the Q-Former with the LM to leverage the LM’s language generation capability, where the molecular representations extracted by the Q-Former serve as 1D soft prompts comprehensible to the LM.
3D Molecule-centric Instruction Tuning fine-tunes the model to follow human instructions on 3D molecule relevant tasks.
- Data Preparation – 3D-MoIT. We construct 3D-MoIT by sourcing data from the PubChem and PubChemQC databases, including both computed molecular properties and descriptive texts that characterize molecule properties in numerical values and free texts, respectively.
- Stage 3: Instruction-based Fine-tuning enhances the ability to follow various instructions and improves the understanding of 3D molecular structures, especially in recognizing 3D-dependent properties
Key dependencies include:
torch==2.0.1
transformers==4.35.0
deepspeed==0.12.2
pytorch-lightning==2.0.7
uni-core==0.0.1See requirements.txt for more detailed requirements.
- 3D-MoIT. Download the dataset from Huggingface-3D-MoIT, and put it under the
./data/directory.
Download following checkpoints from Huggingface-3D-MoLM, and put it under the ./all_checkpoints/ directory.
- 3D-MoLM
- Uni-Mol
- SciBERT
- Llama2-7B
Run inference.ipynb to play with 3D-MoLM.
- Install required conda environment as described in Requirements section
- Download the dataset and required checkpoints as described in Dataset and Checkpoints section.
We provide the outputs of 3D-MoLM on the test set of 3D-MoIT in all_checkpoints/generalist/lightning_logs/version_0/predictions.txt. Run the following script to read it.
python read_generalist_results.py
--file_path 'all_checkpoints/generalist/lightning_logs/version_0/predictions.txt'
--tokenizer_path 'all_checkpoints/llama-2-7b-hf'We share the checkpoint for reproducing results.
bash ./scripts/stage3_test.shStage 1: 3D Molecule-Text Representation Learning
Run the following script for stage 1 pretraining:
bash ./scripts/stage1_pretrain.shStage 2: 3D Molecule-Text Alignment via Generative Learning
Run the following script for stage 2 pretraining:
bash ./scripts/stage2_pretrain.shStage 3: Instruction-based Fine-tuning
Run the following script for instruction tuning:
bash ./scripts/stage3_train.shIf you use our codes or checkpoints, please cite our paper:
@inproceedings{li2024molm,
title={3D-MoLM: Towards 3D Molecule-Text Interpretation in Language Models},
author={Li, Sihang and Liu, Zhiyuan and Luo, Yanchen and Wang, Xiang and He, Xiangnan and Kawaguchi, Kenji and Chua, Tat-Seng and Tian, Qi},
booktitle={ICLR},
year={2024},
url={https://openreview.net/forum?id=xI4yNlkaqh}
}