A hierarchical clustering algorithm for Cytoscape.js.
- Cytoscape.js >= 2.6.12
Download the library:
- via npm:
npm install cytoscape-hierarchical, - via bower:
bower install cytoscape-hierarchical, or - via direct download in the repository.
require() the library as appropriate for your project:
CommonJS:
var cytoscape = require('cytoscape');
var hierarchical = require('cytoscape-hierarchical');
hierarchical( cytoscape ); // register extensionAMD:
require(['cytoscape', 'cytoscape-hierarchical'], function( cytoscape, hierarchical ){
hierarchical( cytoscape ); // register extension
});Plain HTML/JS has the extension registered for you automatically, because no require() is needed.
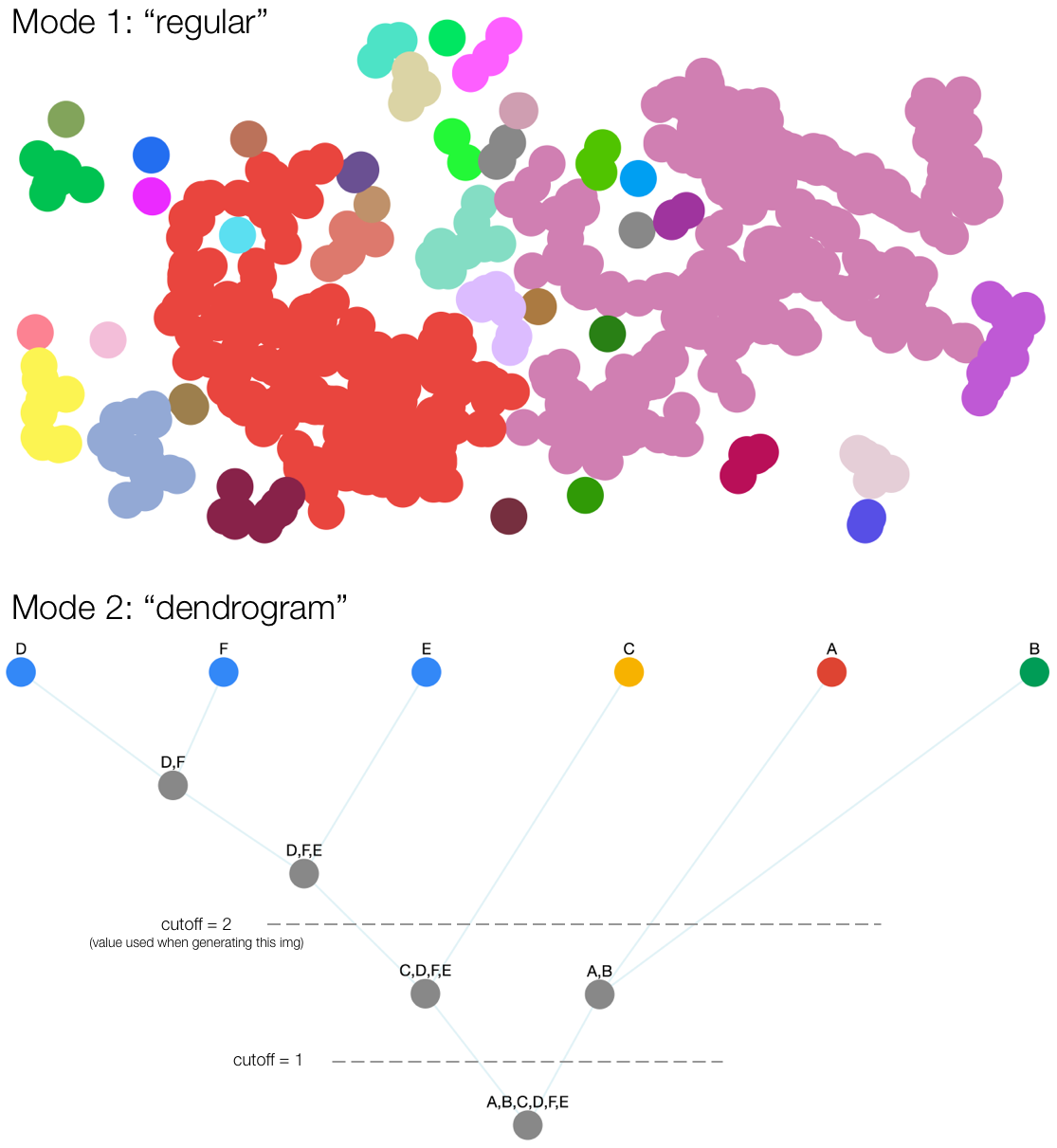
Under the regular mode, the algorithm returns an array of clusters generated from the data set.
One may set the threshold option to specify a stopping point for the algorithm.
When every cluster is more than threshold distance apart, clustering is stopped and the current set of hierarchies is returned.
cy.elements().hierarchical({
mode: "regular", // extension mode
threshold: 25, // stopping criterion that affects granularity (#) of clusters
distance: "euclidean", // distance metric for measuring the distance between two nodes
linkage: "single", // linkage criteria for determining the distance between two clusters
attributes: [ // attributes/features used to group nodes
function(node) {
return node.position('x');
},
function(node) {
return node.position('y');
}
]
});Under the dendrogram mode, the algorithm returns an array of clusters generated from the data set, and generates a dendrogram of the clusters.
One may set the cutoff option to specify the level at which the tree is cut. This option partitions clusters at different precisions.
For example, in the demo img above, setting cutoff = 2 will return the clusters {D,F,E}, {C}, {A}, {B}. Setting cutoff = 1 will return the clusters {D,F,E,C}, {A,B}. Setting cutoff = 0 will return a single cluster containing all the nodes.
Since the dendrogram mode generates many additional nodes and edges in order to render the tree, it might not be performant for large data sets. Thus it is recommended to use regular mode for clustering instead.
cy.elements().hierarchical({
mode: "dendrogram", // extension mode
cutoff: 2, // stopping criterion that affects granularity (#) of clusters
distance: "euclidean", // distance metric for measuring the distance between two nodes
linkage: "single", // linkage criteria for determining the distance between two clusters
attributes: [ // attributes/features used to group nodes
function(node) {
return node.position('x');
},
function(node) {
return node.position('y');
}
]
});demo.html provides working examples of the 2 different modes using separate data sets.
average - the distance between two clusters is an average of the differences between the nodes in the clusters.
single - the distance between clusters is the smallest distance between a node from each cluster.
complete - the distance between clusters is the largest distance between two nodes in the clusters.
This project is set up to automatically be published to npm and bower. To publish:
- Set the version number environment variable:
export VERSION=1.2.3 - Publish:
gulp publish - If publishing to bower for the first time, you'll need to run
bower register cytoscape-hierarchical https://github.com/cytoscape.js-hierarchical.git