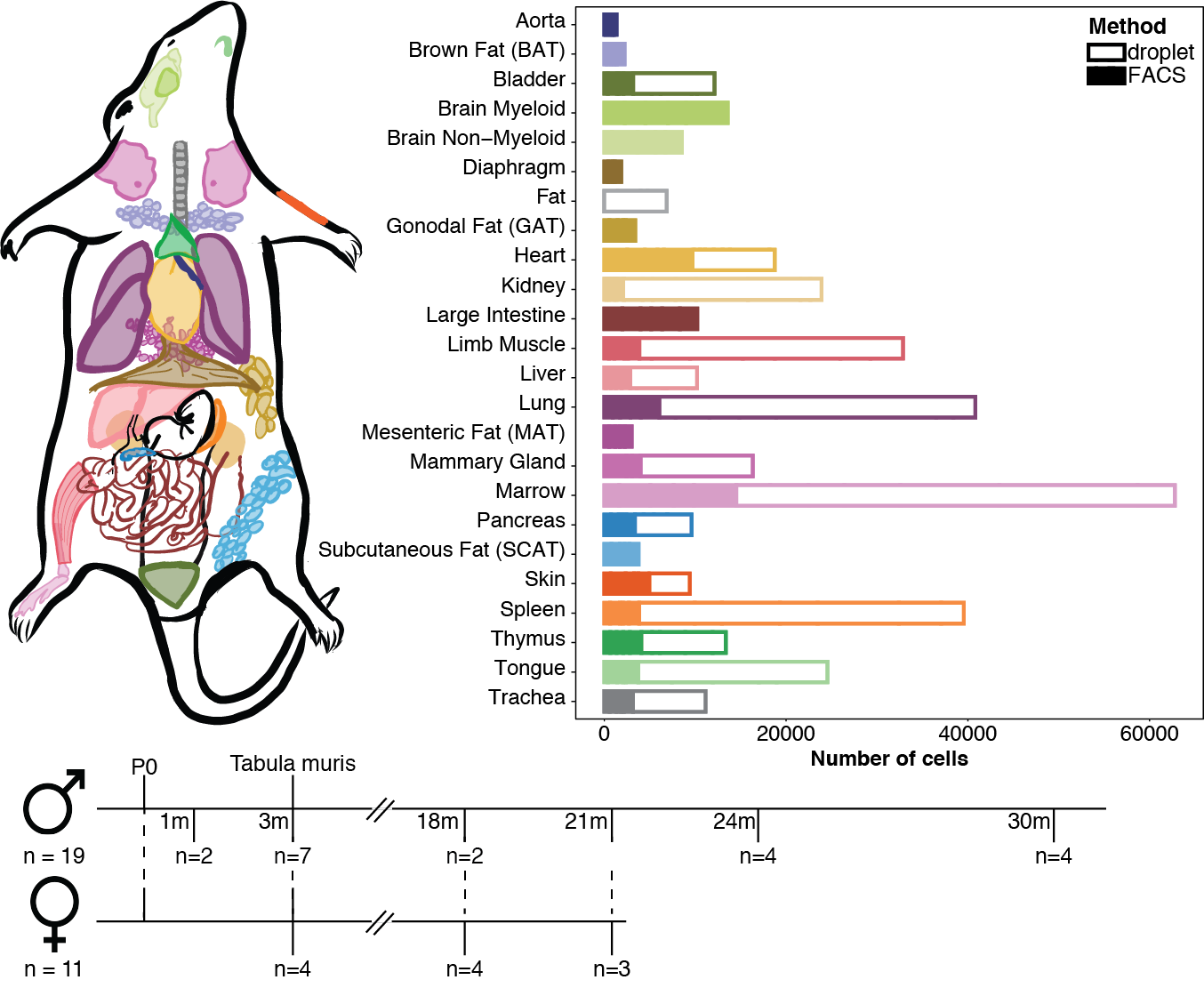
Tabula Muris Senis is a comprehensive resource for the cell biology community which offers a detailed molecular and cell-type specific portrait of aging. You can read our pre-print here!
We view such a cell atlas as an essential companion to the genome: the genome provides a blueprint for the organism but does not explain how genes are used in a cell type specific manner or how the usage of genes changes over the lifetime of the organism. The cell atlas provides a deep characterization of phenotype and physiology which can serve as a reference for understanding many aspects of the cell biological changes that mammals undergo during their lifespan.
For a quick overview checkout the lightining talk slides presented at SciPy2019
This is a resource for the community! The repo is being continously updated with the code used for the publication - let us know your priorities and we will prioritize that code release!
Since October 2019, Tabula Muris Senis data have been made available to all users free of charge. AWS has made the data freely available on Amazon S3 so that anyone can download the resource to perform analysis and advance medical discovery without needing to worry about the cost of storing Tabula Muris Senis data or the time required to download it.
Our ready-to-use with scanpy data is available from figshare: https://figshare.com/projects/Tabula_Muris_Senis/64982
Interactive browsers for the data are available from the Tabula Muris Senis portal using cellxgene and thanks to Max Haeussler and Matt Speir at the UCSC Cell Browser
If you have questions about the data, please create a new Issue.
There are no restrictions on the use of data received from the Chan Zuckerberg Biohub, unless expressly identified prior to or at the time of receipt.