Code repo for AI Through Symbiosis project, accepted for publication by Ambient AI: Multimodal Wearable Sensor Understanding workshop at ICASSP 2023.
Keep track of this repo as we plan to release our picklist video dataset soon!
For a quickstart, you can download the intermediate outputs generated from our dataset and change the config paths to match
Running extract_features.py script to extract features from the videos
# run from ai-through-symbiosis root directory
python3 extract_features.py -v <path to video file>
To visualize features that are being detected (e.g. ArUco markers, hand locations), set DISPLAY_TRUE in
extract_features.py script to True
Next, preprocess the data by running the scripts/data_preprocessing/forward_fill.py and scripts/data_preprocessing/gaussian_convolution.py
scripts. Create a config file by copying scripts/configs/base.yaml and filling in the appropriate paths to folders in your local environment.
To set the configs for the above and subsequent python script calls, use the -c, --config_file argument in the command line call.
Build the docker image that is setup for running The Hidden Markov Model Toolkit (HTK)
cd symbiosis
docker build -t aits:latest .
Enter the container mounting the symbiosis folder
docker run -ti -v $PWD:/tmp/ -w /tmp/ aits /bin/bash
From outside the container, choose and copy the training data and labels from an experiment
cp -R ./ambient_experiments/136-234-aruco1dim/all-data/* ./symbiosis/all-data/
cp -R ./ambient_experiments/136-234-aruco1dim/all-labels/* ./symbiosis/all-labels/
Run n-fold cross validation from within the container (this example has a training/testing split of 80/20 and 20 folds)
./scripts/n_fold.sh -s .8 -d all-data/ -n 20
The output of this script will be a number of fold directories containing the segmentation boundaries of each picklist.
Set the PICKLISTS variable in iterative_improvement_clustering.py to specify the range of picklists numbers to iterate over (script will skip over picklists that are not in the folder)
# run from ai-through-symbiosis root directory
python3 scripts/iterative_improvement_clustering.py
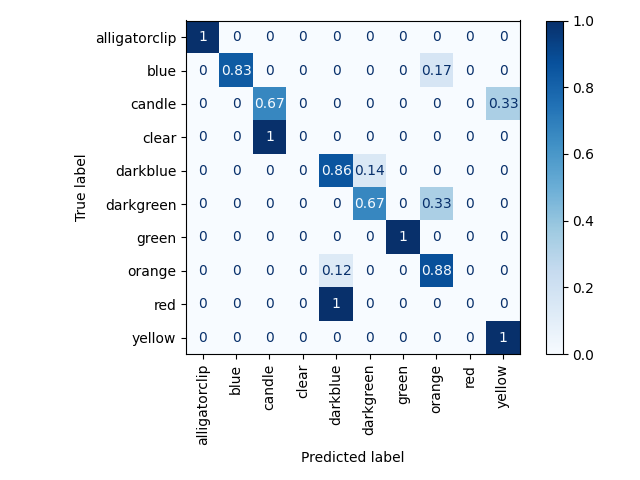
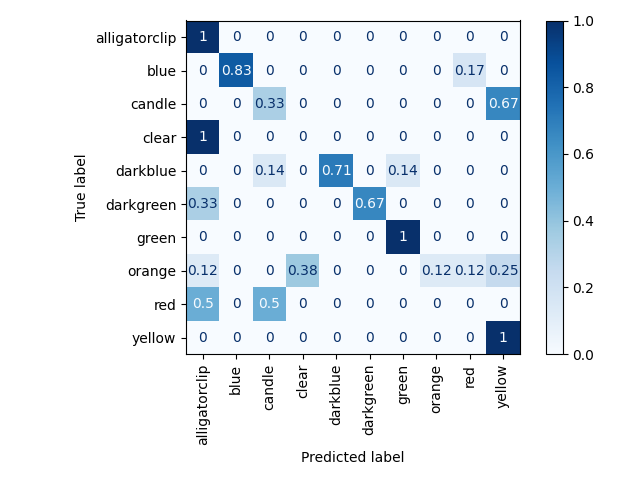
which should give you the predicted labels for the different picklists using the iterative clustering algorithm constrained by the object count set for each picklist. It will also output the hsv bins to object_type_hsv_bins.pkl representing the different object types which serves as our object representation, along with a confusion matrix similar to the results below
After obtaining object_type_hsv_bins.pkl from the previous step, run the object classification script on the relevant picklists by setting the PICKLISTS variable in object_classification.py
# run from ai-through-symbiosis root directory
python3 scripts/object_classification.py
NOTE: If you are using a 30fps video instead of 60fps video, change the
Train set results from clustering (i.e. accuracy of matching labels to pick frame sequences in the train set), which is used to
label the weakly supervised action sets and obtain object representations
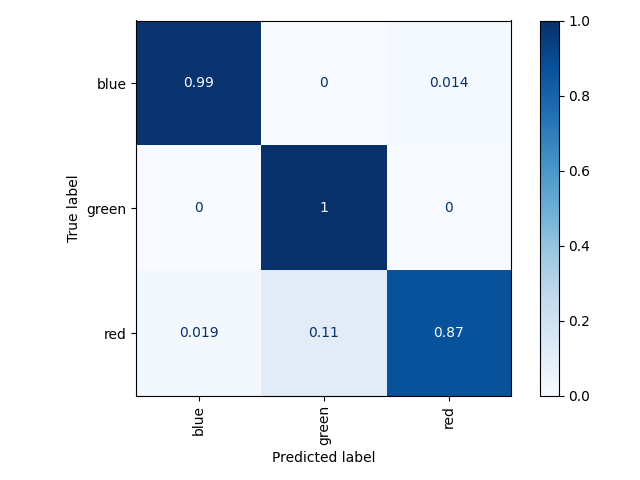
Test set results on three object test set (restricting output predictions to three objects):
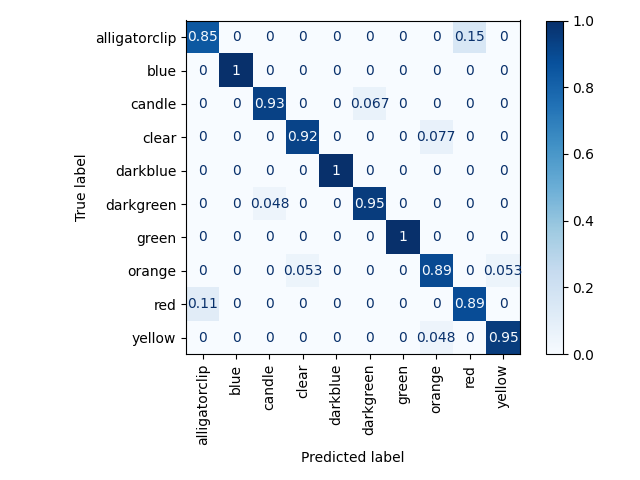
Test set results on ten object test set (constrained to set of objects that appeared in the picklist):