SCORPIUS
SCORPIUS an unsupervised approach for inferring linear developmental chronologies from single-cell RNA sequencing data. In comparison to similar approaches, it has three main advantages:
-
It accurately reconstructs linear dynamic processes. The performance was evaluated using a quantitative evaluation pipeline and ten single-cell RNA sequencing datasets.
-
It automatically identifies marker genes, speeding up knowledge discovery.
-
It is fully unsupervised. Prior knowledge of the relevant marker genes or cellular states of individual cells is not required.
News:
-
See
news(package = "SCORPIUS")for a full list of changes to the package. -
A preprint is available on bioRxiv. Run
citation("SCORPIUS")to obtain the corresponding citation information. -
Check out our review on Trajectory Inference methods!
Installing SCORPIUS
You can install:
-
the latest released version from CRAN with
install.packages("SCORPIUS") -
the latest development version from GitHub with
devtools::install_github("rcannood/SCORPIUS", build_vignettes = TRUE)
If you encounter a bug, please file a minimal reproducible example on the issues page.
Learning SCORPIUS
To get started, read the introductory example below, or read one of the vignettes containing more elaborate examples:
- Investigating dendritic cell maturation in dendritic cell
progenitors:
vignette("ginhoux", package="SCORPIUS") - Trajectory inference from simulated
data:
vignette("simulated-data", package="SCORPIUS")
Introductory example
This section describes the main workflow of SCORPIUS without going in depth in the R code. For a more detailed explanation, see the vignettes listed below.
To start using SCORPIUS, simply write:
library(SCORPIUS)The ginhoux dataset (See Schlitzer et al. 2015) contains 248 dendritic
cell progenitors in one of three cellular cellular states: MDP, CDP or
PreDC. Note that this is a reduced version of the dataset, for packaging
reasons. See ?ginhoux for more info.
data(ginhoux)
expression <- ginhoux$expression
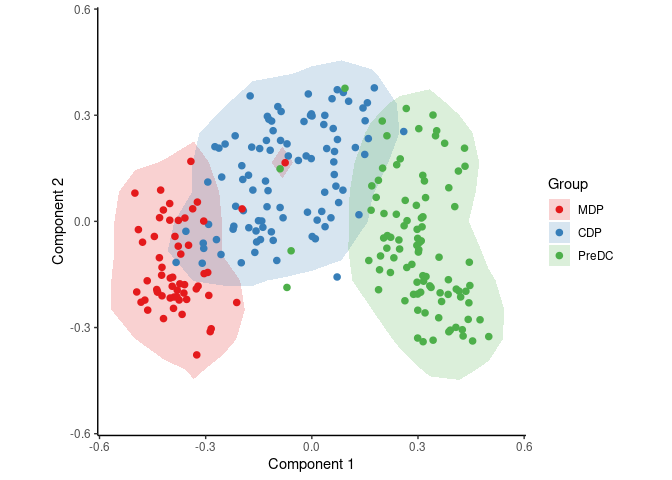
group_name <- ginhoux$sample_info$group_nameWith the following code, SCORPIUS reduces the dimensionality of the dataset and provides a visual overview of the dataset. In this plot, cells that are similar in terms of expression values will be placed closer together than cells with dissimilar expression values.
space <- reduce_dimensionality(expression, "spearman")
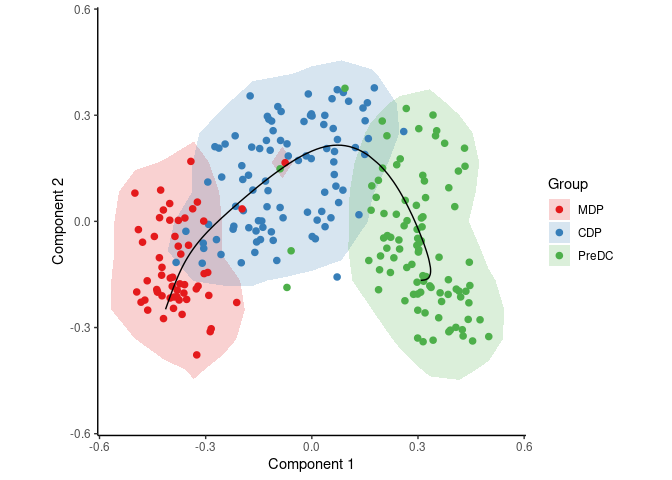
draw_trajectory_plot(space, group_name, contour = TRUE)To infer and visualise a trajectory through these cells, run:
traj <- infer_trajectory(space)
draw_trajectory_plot(space, group_name, traj$path, contour = TRUE)To identify candidate marker genes, run:
# warning: setting num_permutations to 10 requires a long time (~30min) to run!
# set it to 0 and define a manual cutoff for the genes (e.g. top 200) for a much shorter execution time.
gimp <- gene_importances(
expression,
traj$time,
num_permutations = 10,
num_threads = 8,
ntree = 10000,
ntree_perm = 1000
) To select the most important genes and scale its expression, run:
gimp$qvalue <- p.adjust(gimp$pvalue, "BH", length(gimp$pvalue))
gene_sel <- gimp$gene[gimp$qvalue < .05]
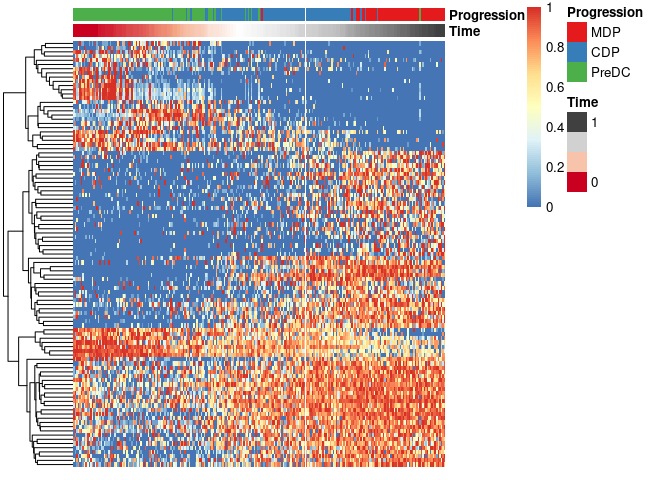
expr_sel <- scale_quantile(expression[,gene_sel])To visualise the expression of the selected genes, use the
draw_trajectory_heatmap function.
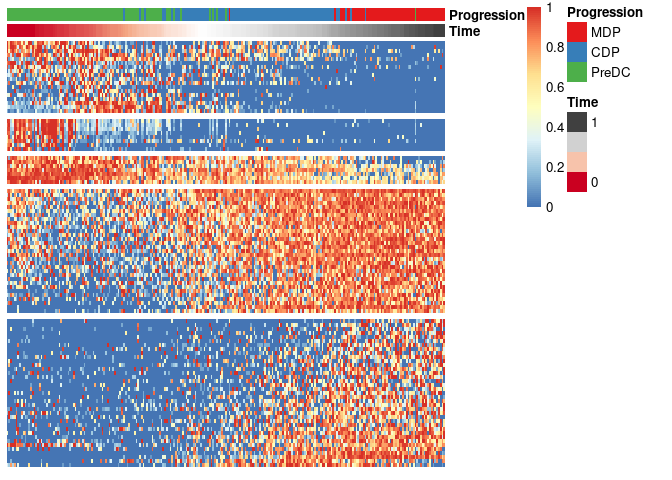
draw_trajectory_heatmap(expr_sel, traj$time, group_name)Finally, these genes can also be grouped into modules as follows:
modules <- extract_modules(scale_quantile(expr_sel), traj$time, verbose = F)
draw_trajectory_heatmap(expr_sel, traj$time, group_name, modules)Latest changes
Check out news(package = "SCORPIUS") or NEWS.md for a full
list of changes.
Recent changes in SCORPIUS 1.0.5
Major change
- Added a
ti_scorpius()wrapper to SCORPIUS.
Minor change
-
Use
RANN::nn2()instead of own nearest neighbour functions. -
Remove deprecated functions.
-
Use
lmdsinstead ofdyndimred.
Recent changes in SCORPIUS 1.0.4 (07-08-2019)
Minor changes
- Added extra customisation parameters to
draw_trajectory_plot()anddraw_trajectory_heatmap().
Optimisation
-
Fixed internal function
check_numeric_matrix()such that it does not run for ages when applied to a large sparse matrix. -
Minor improvement in
infer_initial_trajectory()when calculating the distance from points to along candidate segments.
References
Schlitzer, Andreas, V Sivakamasundari, Jinmiao Chen, Hermi Rizal Bin Sumatoh, Jaring Schreuder, Josephine Lum, Benoit Malleret, et al. 2015. “Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow.” Nature Immunology 16 (7): 718–26. https://doi.org/10.1038/ni.3200.