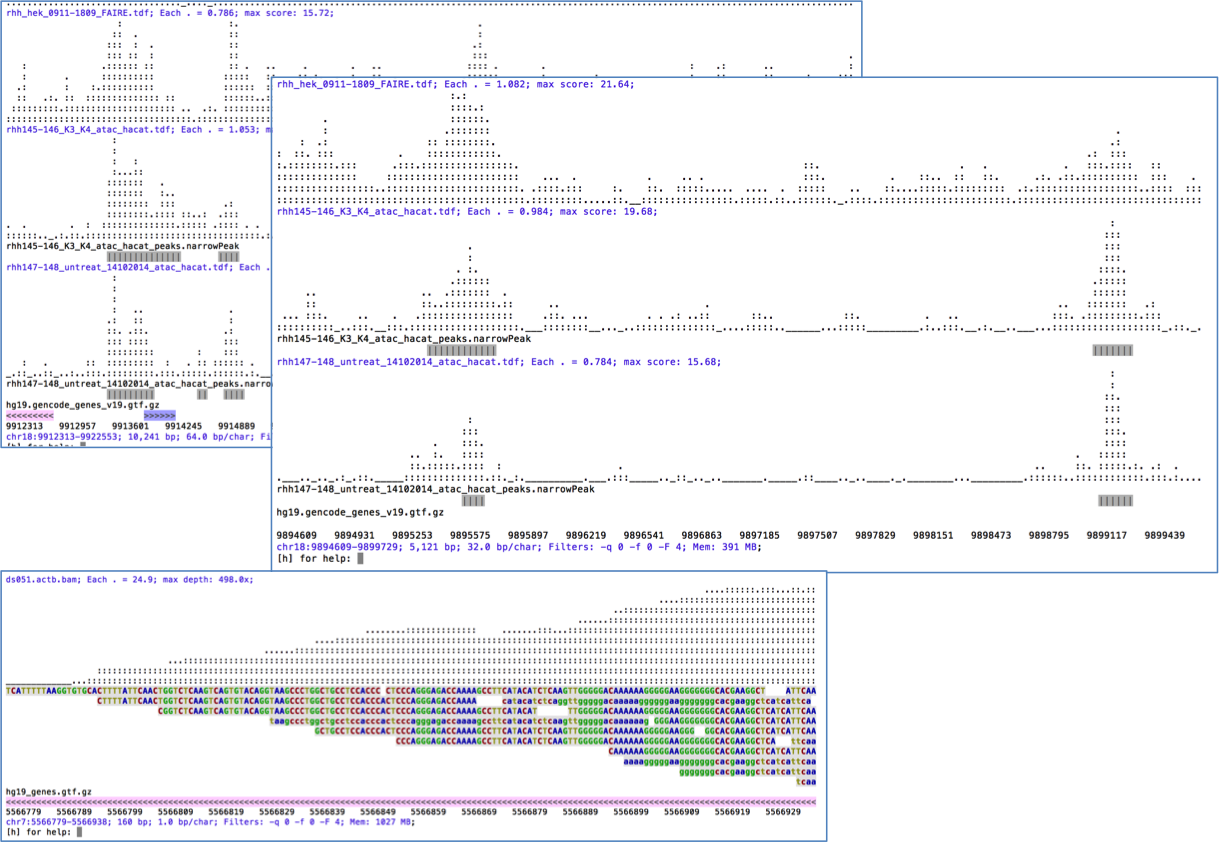
Text Only Genome Viewer!
- Description
- Requirements and Installation
- Usage examples
- Supported input
- Genome option
- Formatting of reads and features
- Saving screenshots
- Tips gotchas and miscellanea
- Interactive commands
- Credits
Description
ASCIIGenome is a command-line genome browser running from terminal window and solely based on
ASCII characters. Since ASCIIGenome does not require a graphical interface it is particularly
useful for quickly visualizing genomic data on remote servers. The idea is to make ASCIIGenome
the Vim of genome viewers.
As far as I know, the closest program to ASCIIGenome is samtools tview but
ASCIIGenome offers much more flexibility, similar to popular GUI viewers like IGV.
Some key features:
- Command line input and interaction, no graphical interface, minimal installation and requirements
- Can load multiple files in various formats
- Can access remote files via URL or ftp address
- Easy navigation and searching of features and sequence motifs and filtering options
- Support for BS-Seq alignment
Requirements and Installation
Installation quick start
In the commands below replace version number with the latest from releases:
wget https://github.com/dariober/ASCIIGenome/releases/download/v0.1.0/ASCIIGenome-0.2.0.zip
unzip ASCIIGenome-0.2.0.zip
cd ASCIIGenome-0.2.0/
chmod a+x ASCIIGenome
cp ASCIIGenome.jar /usr/local/bin/ # Or ~/bin/
cp ASCIIGenome /usr/local/bin/ # Or ~/bin/
Installation through Homebrew
ASCIIGenome can also be installed through brew / Linux Brew, although it is still not an official package:
brew install https://raw.githubusercontent.com/dariober/ASCIIGenome/master/install/brew/asciigenome.rb
A little more detail
ASCIIGenome.jar requires Java 1.7+ and this should be the only requirement. There is virtually no installation needed as ASCIIGenome is pure Java and should work on most (all?) platforms. Download the zip file ASCIIGenome-x.x.x.zip from releases, unzip it and execute the jar file with
java -jar /path/to/ASCIIGenome.jar --help
To avoid typing java -jar ... every time, you can put both the helper
script ASCIIGenome and the jar file ASCIIGenome.jar in the same directory in your PATH and execute with:
ASCIIGenome [options]
Note the helper is a bash script. To set the amount of memory available to java use the -Xmx option as e.g. java -Xmx1500m -jar ....
If for some reason the text formatting misbehaves, disable it with the -nf option. I have developed
ASCIIGenome on MacOS, Ubuntu and CentOS with bash 4.1, white colour background.
Usage examples
These are just some functionalities to give an idea behind ASCIIGenome.
Minimal example
Open an indexed bam file, as simple as:
ASCIIGenome aln.bam
Open with a reference genome (reference must be indexed, see Supported input):
ASCIIGenome -fa genome.fa aln.bam
Open and browse
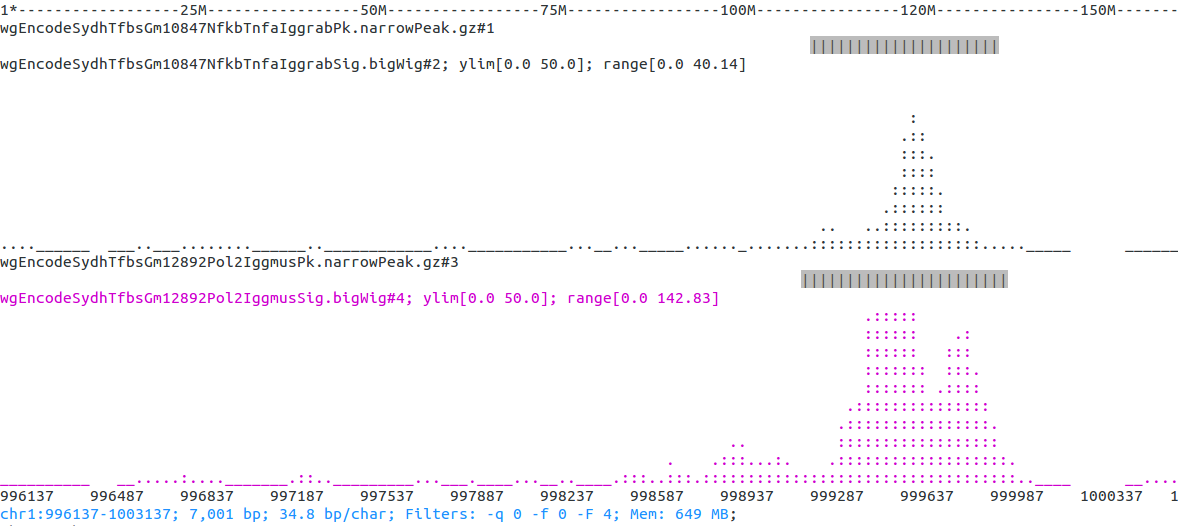
Open some peak and bigWig files from ENCODE. Note that opening remote bigwig files is a little slow (IGV seems equally slow) and it might not work with some proxy settings (see also issue#6):
encode=http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeSydhTfbs
ASCIIGenome -g hg19 \
$encode/wgEncodeSydhTfbsGm10847NfkbTnfaIggrabPk.narrowPeak.gz \
$encode/wgEncodeSydhTfbsGm10847NfkbTnfaIggrabSig.bigWig \
$encode/wgEncodeSydhTfbsGm12892Pol2IggmusPk.narrowPeak.gz \
$encode/wgEncodeSydhTfbsGm12892Pol2IggmusSig.bigWig
Find the first feature on the first file, then change colour of one of the tracks. Reset y axes to span 0 to 50, finally save as png to default file name:
[h] for help: next #1
[h] for help: colorTrack magenta wgEncodeSydhTfbsGm12892Pol2IggmusSig
[h] for help: ylim 0 50
[h] for help: save .png
Result on terminal screen should look like this:
Saved file is chr1_996137-1003137.png (currently the png output doesn't include colours though).
Finding & filtering stuff
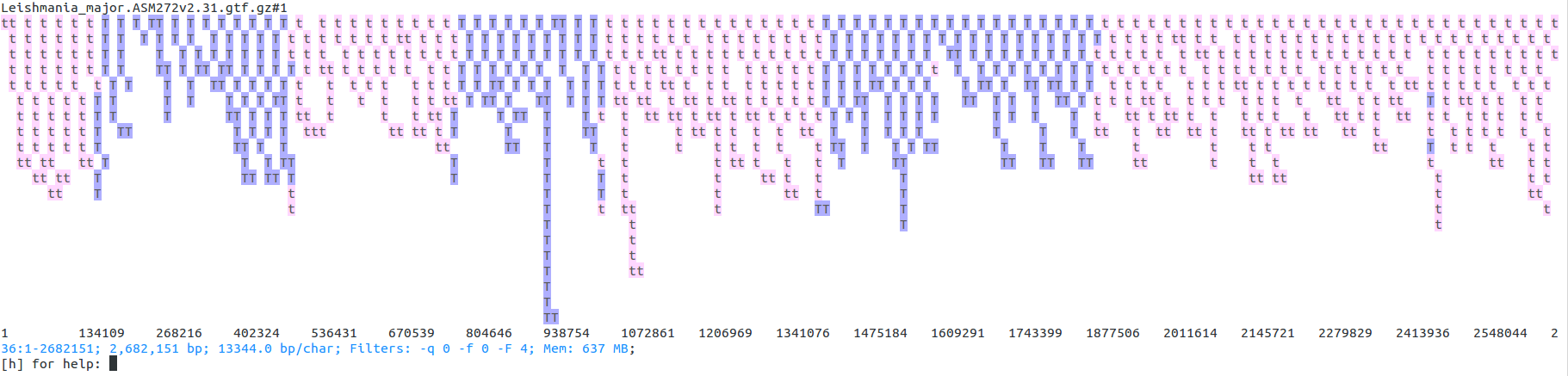
Once started, ASCIIGenome makes it easy to browse the genome. The picture below shows the distribution of transcripts on chromosome 36 of Leishmania major. It is clearly visible how transcripts in Leishmania tend to be grouped in blocks transcribed from the same direction (blue: forward strand, pink: reverse strand). Note how overlapping features are stacked on top of each other.
This screenshot has been produced by first loading the L. major GTF file:
ASCIIGenome ftp://ftp.ensemblgenomes.org/pub/release-31/protists/gtf/leishmania_major/Leishmania_major.ASM272v2.31.gtf.gz
At command prompt issue the following commands:
[h] for help: goto 36:1-2682151
[h] for help: filter \ttranscript\t
[h] for help: trackHeight 100
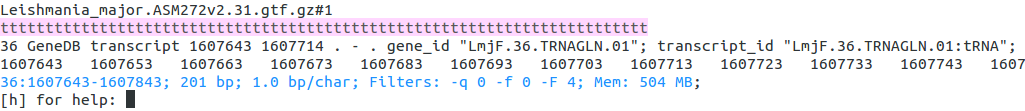
Now return to the start of the chromosome and find the first feature containing LmjF.36.TRNAGLN.01, print it to screen:
[h] for help: 1
[h] for help: find_first LmjF.36.TRNAGLN.01
[h] for help: print
Now showing:
Chaining commands
Commands need not to be executed one at a time but can be chained with the && operator (like in Bash). This is more
convenient than executing commands one by one and it is also faster as tracks are processed only once. For example, the example above
could be executed in one pass as
goto 36:1-2682151 && filter \ttranscript\t && trackHeight 100
In addition, the same could be achieved at the start via the --exec/-x option:
ASCIIGenome -x 'goto 36:1-2682151 && filter \ttranscript\t && trackHeight 100' \
ftp://ftp.ensemblgenomes.org/pub/release-31/protists/gtf/leishmania_major/Leishmania_major.ASM272v2.31.gtf.gz
Note that if the first option passed to -exec/-x starts with - you need to add a space between
the opening quote and the option itself. For example do ASCIIGenome -x ' -F 16' ... instead of
ASCIIGenome -x '-F 16' ....
Supported input
File name extensions matter as file types are usually recognized by their extension in case insensitive mode.
- bam files should be sorted and indexed, e.g. with
samtools sortandsamtools index. Paths to remote URLs are supported but painfully slow (IGV seems to suffer of the same issue). - bigWig recognized by extension
.bwor.bigWig. Remote URLs supported. - bedGraph recognized by extension
.bedGraphor.bedgraph - bed, gtf, gff recognized by respective extensions. Remote URLs supported.
- tdf This is very useful for quickly displaying very large intervals like tens of megabases or entire chromosomes see tdf
- vcf Supported but not too sophisticated representation. URL should be supported but it appears ftp from 1000genomes doesn't work (same for IGV).
- All other extensions (e.g. txt, narrowPeak) will be treated as bed files, provided the format is actually bed!
All plain text formats (bed, bedgraph, etc) can be read as gzipped and there is no need to decompress them.
Bedgraph files should be sorted by position, a sort -k1,1 -k2,2n will do. Unindexed bedGraph files are first bgzipped and indexed to temporary files which are deleted on exit. This can take time for large files so consider creating the index once for all with tabix, e.g.
bgzip my.bedgraph &&
tabix -p bed my.bedgraph.gz
Bed & gtf file are not required to be sorted or index but in this case they are loaded in memory. To save memory and time for large files you can again index them as above. Loading in memory is typically fast for files of up to ~1/2 million records.
For input format specs see also UCSC format and for guidelines on the choice of format see IGV recommendations.
Fasta reference: The reference sequence should be uncompressed and indexed, with e.g. samtools faidx:
samtools faidx genome.fa
Notable formats currently not supported: cram, bigBed.
bigBed files can be converted to bgzip format with bigBedToBed from
UCSC utilities and then indexed with tabix. For example:
bigBedToBed input.bb /dev/stdout/ | bgzip > input.bed.gz
tabix -p bed input.bed.gz
Genome option
An optional genome file can be passed to option -g/--genome to give a set of allowed sequences and their sizes so that browsing is constrained to the real genomic space.
The genome file is also used to represent the position of the current window on the chromosome, which is handy to navigate around.
There are three ways to pass a genome file:
-
A tag identifying a built-in genome, e.g. hg19. See genomes for available genomes
-
A local file, tab separated with columns chromosome name and length. See genomes for examples.
-
A bam file with suitable header.
Note that if the input list of files contains a bam file, the --genome option is effectively ignored as the genome dictionary is extracted from the bam header.
Formatting of reads and features
When aligned reads are show at single base resolution, read bases follow the same convention as samtools:
Upper case letters and . for read align to forward strand, lower case and , otherwise; second-in-pair reads are underlined;
grey-shaded reads have mapping quality of <=5.
GTF/GFF features on are coded according to the feature column as below. For forward strand features the colour blue and upper case is used, for reverse strand the colour is pink the case is lower. Features with no strand information are in grey.
| Feature | Symbol |
|---|---|
| exon | E |
| cds | C |
| start_codon | A |
| stop_codon | Z |
| utr | U |
| 3utr | U |
| 5utr | W |
| gene | G |
| transcript | T |
| mrna | M |
| trna | X |
| rrna | R |
| mirna | I |
| ncrna | L |
| lncrna | L |
| sirna | S |
| pirna | P |
| snorna | O |
If available, the feature name is shown on the feature itself.
The feature name has a trailing underscore to separate it from the rest of the feature representation. The
last character of the feature is always the feature type. For example, the feature named myGene appears as:
myGene_EEEEEEEEE ## Enough space for the full name
myGenE ## Not enough space, name truncated and last char is E
For BED features, name is taken from column 4, if available. Default for GTF/GFF is to take name from attribute
Name, if absent try: ID, transcript_name, transcript_id, gene_id, gene_name.
To choose an attribute see command gffNameAttr.
Read coverage tracks at single base resolution show the consensus sequence obtained from the underlying reads. If
the reference fasta file is present the = symbol is used to denote a match. Heterozygote bases or variants are shown
using the iupac ambiguity codes for up to two variants (N otherwise).
Variants are called with a not-too-sophisticated heuristics: Only base qualities >= 20 are considered, an alternative allele
is called if supported by at least 3 reads and makes up at least 1% of the total reads. The first and second allele must make at least
98% of the total reads otherwise the base is N (see PileupLocus.getConsensus() for exact implementation). Insertion/deletions
are currently not considered.
Saving screenshots
Screenshots can be saved to file with the commands save. Output format is either ASCII text or
png, depending on file name extension. For example:
[h] for help: save mygene.txt ## Save to mygene.txt as text
[h] for help: save ## Save to chrom_start-end.txt as text
[h] for help: save .png ## Save to chrom_start-end.png as png
[h] for help: save mygene.png ## Save to mygene.png as png
Without arguments, save writes to file named after the current genomic position e.g.
chr1_1000-2000.txt. The ANSI formatting (i.e. colours) is stripped before saving so that files
can be viewed on any text editor (use a monospace font like courier).
Tips gotchas and miscellanea
-
Performance Alignment files are typically accessed very quickly but
ASCIIGenomebecomes slow when the window size grows above a few hundreds of kilobases. Annotation files (bed, gff, gtf) are loaded in memory unless they are indexed withtabix. -
Regular expression Use the
(?i)modifier to match in case insensitve mode, e.g. '(?i).actb.' -
When displaying bam files,
ASCIGenomeis hardcoded to disable the coverage and read tracks if the window size is >100,000 bp. This is to prevent the browsing to become horribly slow. To display such large windows consider bigWig or tdf file format. -
When opening bam files, the first chromosome is often the mitochondrial chromosome chrM (or chrMT) which often has very high read depth (say 10,000x). This can make the opening slow. Consider using the
-roption in these cases. E.g.ASCIIGenome -r chr1 file1.bam file2.bam ...
Interactive commands
The description of each interactive commands is here
As there is no GUI, everything is handled thorough command line. Once ASCIIGenome is started enter
a command and press ENTER to execute.
Some features of Unix console are enabled:
- Arrow keys UP and DOWN scroll previous commands.
- TAB auto-completes commands.
- ENTER without any argument repeats the previous command.
Examples:
[h] for help: ff <ENTER> ## Move forward
[h] for help: <ENTER> ## Move forward again...
[h] for help: <ENTER> ## ... and again
[h] for help: col <TAB> ## Is expanded to colorTrack
[h] for help: <ARROW UP> ## Shows previous command
[h] for help: h <ENTER> ## Show help.
When track names are passed as arguments, it is not necessary to give the full name as
partial matching is enabled. This is handy since track names have an ID appended as suffix which can
be used in place of the full name, e.g. next myLongfileName.bed#1 can be also typed as next #1.
Credits
- Bam processing is mostly done with the samtools/htsjdk library.
- Bigwig and tdf are processed with classes from IGV source code.
- Block compression and indexing done using jvarkit.
- Brew installation thanks to dalloliogm.