This is a tool for simulating next-generation sequencing data from a hierarchical sampling arrangement with single-cell and bulk samples.
- Clone the repository to your computer.
- Inside the repository, run
make allto build the SCSIM docker image and run the example.
- docker
You can check your docker installation with $ docker run hello-world.
If you get an error that you don't have permissions to run docker, the official documentation has the way to fix that error https://docs.docker.com/install/linux/linux-postinstall/#manage-docker-as-a-non-root-user.
- Create the docker group.
$ sudo groupadd docker- Add your user to the docker group.
$ sudo usermod -aG docker $USER- Log out and log back in so that your group membership is re-evaluated (you may need a restart).
- Verify that you can run docker commands without sudo.
$ docker run hello-worldIf you are testing or running this software in a virtualbox environment, make sure you allocate at least 20Gb of disk space and 8Gb of RAM.
You can run the project directly from a docker container by running make all which runs make build and make example.
The conda environment can be installed using make env. But, the example needs to run monovar which requires a different version of python. So, it's best to build the example using the make all command.
The scsim docker image must be built before running the example.
- Navigate the
exampledirectory in the repository. - Run
make all
A docker container will be created and the example simulation will be run in the container.
The simulation output will appear in the example folder because a shared volume is created in the container that connects to the example folder.
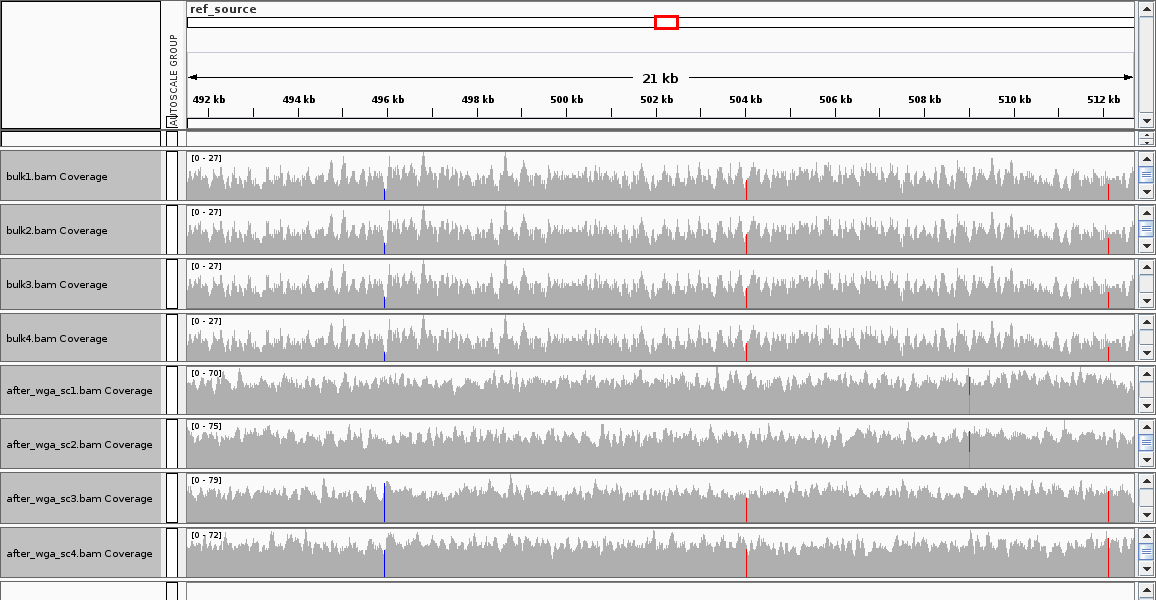
The example/test folder contains expected results: a csv file showing the actual SNV locations across the three prototypes and txt files showing where bcftools and monovar called SNVs.
An IGV plot of the example output is shows shares mutations in bulk and single-cell samples.
If you'd like to contribute please fork the repository and submit a pull request. Or if you find an issue, add an issue to the github repository.
- Collin Giguere
- Harsh Dubey
- Vishal Sarsani
- Hachem Saddiki
- Shai He
- Patrick Flaherty
MIT (see LICENSE.md)